Abstract
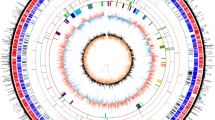
To date, several actinomycete genomes have been completed and annotated. Among them, Streptomyces microorganisms are of major pharmaceutical interest because they are a rich source of numerous secondary metabolites. S. avermitilis is an industrial microorganism used for the production of an anthelmintic agent, avermectin, which is a commercially important antiparasitic agent in human and veterinary medicine, and agricultural pesticides. Genome analysis of S. avermitilis provides significant information for not only industrial applications but also understanding the features of this genus. On genome mining of S. avermitilis, the microorganism has been found to harbor at least 38 secondary metabolic gene clusters and 46 insertion sequence (IS)-like sequences on the genome, which have not been searched so far. A significant use of the genome data of Streptomyces microorganisms is the construction of a versatile host for heterologous expression of exogenous biosynthetic gene clusters by genetic engineering. Since S. avermitilis is used as an industrial microorganism, the microorganism is already optimized for the efficient supply of primary metabolic precursors and biochemical energy to support multistep biosynthesis. The feasibility of large-deletion mutants of S. avermitilis has been confirmed by heterologous expression of more than 20 exogenous biosynthetic gene clusters.







Similar content being viewed by others
References
Barbe V, Bouzon M, Mangenot S, Badet B, Poulain J, Segurens B, Vallenet D, Marlière P, Weissenbach J (2011) Complete genome sequence of Streptomyces cattleya NRRL 8057, a producer of antibiotics and fluorometabolites. J Bacteriol 193:5055–5056
Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, Bateman A, Brown S, Chandra G, Chen CW, Collins M, Cronin A, Fraser A, Goble A, Hidalgo J, Hornsby T, Howarth S, Huang CH, Kieser T, Larke L, Murphy L, Oliver K, O’Neil S, Rabbinowitsch E, Rajandream MA, Rutherford K, Rutter S, Seeger K, Saunders D, Sharp S, Squares R, Squares S, Taylor K, Warren T, Wietzorrek A, Woodward J, Barrell BG, Parkhill J, Hopwood DA (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147
Bignell DRD, Seipke RF, Huguet-Tapia JC, Chambers AH, Parry RJ, Loria R (2010) Streptomyces scabies 87-22 contains a coronafacic acid-like biosynthetic cluster that contributes to plant–microbe interactions. Mol Plant Microbe Interact 23:161–175
Burg RW, Miller BM, Baker EE, Birnbaum J, Currie SA, Hartman R, Kong YL, Monaghan RL, Olson G, Putter I, Tunac JB, Wallick H, Stapley EO, Oiwa R, Omura S (1979) Avermectins, new family of potent anthelmintic agents: producing organism and fermentation. Antimicrob Agents Chemother 15:361–367
Cane DE, He X, Kobayashi S, Omura S, Ikeda H (2006) Geosmin biosynthesis in Streptomyces avermitilis. J Antibiot 59:471–479
Cane DE, You Z, Omura S, Ikeda H (2007) Pentalenolactone biosynthesis. Molecular cloning and assignment of biochemical function to PtlF, a short-chain dehydrogenase from Streptomyces avermitilis, and identification of a new biosynthetic intermediate. Arch Biochem Biophys 459:233–240
Cánovas D, Vargas C, Calderon MI, Ventosa A, Nieto JJ (1998) Characterization of the genes for the biosynthesis of the compatible solute ectoine in the moderately halophilic bacterium Halomonas elongata DSM 3043. Syst Appl Microbiol 21:487–497
Carter GT, Nietsche JA, Hertz MR, Williams DR, Siegel MM, Morton GO, James JC, Borders DB (1988) LL-F28249 antibiotic complex: a new family of antiparasitic macrocyclic lactones: isolation, characterization and structures of LL-F28249 α, β, γ, λ. J Antibiot 41:519–529
Chou WKW, Fanizza I, Uchiyama T, Komatsu M, Ikeda H, Cane DE (2010) Genome mining in Streptomyces avermitilis: cloning and characterization of SAV_76, the synthase for a new sesquiterpene, avermitilol. J Am Chem Soc 132:8850–8851
Chung ST (1987) Tn4556, a 6.8-kilobase-pair transposable element of Streptomyces fradiae. J Bacteriol 169:4436–4441
Denoya CD, Skinner DD, Morgenstern MR (1994) A Streptomyces avermitilis gene encoding a 4-hydroxyphenylpyruvic acid dioxygenase-like protein that directs the production of homogentisic acid and an ochronotic pigment in Escherichia coli. J Bacteriol 176:5312–5319
Feng Z, Wang L, Rajski SR, Xu Z, Coeffet-LeGal MF, Shen B (2009) Engineered production of iso-migrastatin in heterologous Streptomyces hosts. Bioorg Med Chem 17:2147–2153
Hsiao NH, Nakayama S, Merlo ME, Vries M, Bunet R, Kitani S, Nihira T, Takano T (2009) Analysis of two additional signaling molecules in Streptomyces coelicolor and the development of a butyrolactone-specific reporter system. Chem Biol 16:951–960
Ikeda H, Kotaki H, Omura S (1987) Genetic studies of avermectin biosynthesis in Streptomyces avermitilis. J Bacteriol 169:5615–5621
Ikeda H, Takada Y, Pang CH, Tanaka H, Omura S (1993) Transposon mutagenesis by Tn4560 and applications with avermectin-producing Streptomyces avermitilis. J Bacteriol 175:2077–2082
Ikeda H, Nonomiya T, Usami M, Ohta T, Omura S (1999) Organization of the biosynthetic gene cluster for the polyketide anthelmintic macrolide avermectin in Streptomyces avermitilis. Proc Natl Acad Sci U S A 96:9509–9514
Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S (2003) Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol 21:526–531
Inbar L, Lapidot A (1988) The structure and biosynthesis of new tetrahydropyrimidine derivatives in actinomycin D producer Streptomyces parvulus. J Biol Chem 263:16014–16022
Inokoshi J, Matsuhama M, Miyake M, Ikeda H, Tomoda H (2012) Molecular cloning of the gene cluster for lariatin biosynthesis of Rhodococcus jostii K01-B0171. Appl Microb Biotechnol 95:451–460
Jiang J, Tetzlaff CN, Takamatsu S, Iwatsuki M, Komatsu M, Ikeda H, Cane DE (2009) Genome mining in Streptomyces avermitilis: a biochemical Baeyer-Villiger reaction and discovery of a new branch of the pentalenolactone family tree. Biochemistry 48:6431–6440
Kitani S, Miyamoto KT, Takamatsu S, Herawati E, Iguchi H, Nishitomi K, Uchida M, Nagamitsu T, Omura S, Ikeda H, Nihira T (2011) Avenolide, a Streptomyces hormone controlling antibiotic production in Streptomyces avermitilis. Proc Natl Acad Sci U S A 108:16410–16415
Komatsu M, Tsuda M, Omura S, Oikawa H, Ikeda H (2008) Identification and functional analysis of genes controlling biosynthesis of 2-methylisoborneol. Proc Natl Acad Sci U S A 105:7422–7427
Komatsu M, Uchiyama T, Omura S, Cane DE, Ikeda H (2010) Genome-minimized Streptomyces host for the heterologous expression of secondary metabolism. Proc Natl Acad Sci U S A 107:2646–2651
Komatsu M, Komatsu K, Koiwai H, Yamada Y, Kozone I, Izumikawa M, Hashimoto J, Takagi M, Omura S, Shin-ya K, Cane DE, Ikeda H (2013) Engineered Streptomyces avermitilis host for heterologous expression of biosynthetic gene cluster for secondary metabolites. ACS Syn Biol 2:384–396
Kuhlmann AU, Bremer E (2002) Osmotically regulated synthesis of the compatible solute ectoine in Bacillus pasteurii and related Bacillus spp. Appl Env Microbiol 68:772–783
Lamb DC, Ikeda Nelson DR, Ishikawa J, Skaug T, Jackson C, Omura S, Waterman MR, Kelly SL (2003) Cytochrome P450 complement of the avermectin-producer Streptomyces avermitilis and comparison to that of Streptomyces coelicolor A3(2). Biochem Biophys Res Commun 307:610–619
Lautru S, Deeth RJ, Bailey LM, Challis GL (2005) Discovery of a new peptide natural product by Streptomyces coelicolor genome mining. Nat Chem Biol 1:265–269
Lin X, Cane DE (2009) Biosynthesis of sesquiterpene antibiotic albaflavenone in Streptomyces coelicolor. Mechanism and stereochemistry of the enzymatic formation of epi-isozizaene. J Am Chem Soc 131:6332–6333
Louis P, Galinski EA (1997) Characterization of genes for the biosynthesis of the compatible solute ectoine from Marinococcus halophilus and osmoregulated expression in Escherichia coli. Microbiology 143:1141–1149
Malin G, Lapidot A (1996) Induction of synthesis of tetrahydropyrimidine derivatives in Streptomyces strains and their effect on Escherichia coli in response to osmotic and heat stress. J Bactriol 178:385–395
Martinez A, Kolvek SJ, Yip CLT, Hopke J, Brown KA, MacNeil IA, Osburne MS (2004) Genetically modified bacterial strains and novel bacterial artificial chromosome shuttle vectors for constructing environmental libraries and detecting heterologous natural products in multiple expression hosts. Appl Environ Microbiol 70:2452–2463
Medema MH, Trefzer A, Kovalchuk A, Berg M, Müller U, Heijne W, Wu L, Alam MT, Ronning CM, Nierman WC, Bovenberg RAL, Breitling R, Takano E (2010) The sequence of a 1.8-Mb bacterial linear plasmid reveals a rich evolutionary reservoir of secondary metabolic pathways. Genome Biol Evol 2:212–224
Nett M, Ikeda H, Moore BS (2009) Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep 26:1326–1384
Ohnishi Y, Ishikawa J, Hara H, Suzuki H, Ikenoya M, Ikeda H, Yamashita A, Hattori M, Horinouchi S (2008) Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J Bacteriol 190:4050–4060
Omura S, Ikeda H, Ishikawa J, Hanamoto A, Takahashi C, Shinose M, Takahashi Y, Horikawa H, Nakazawa H, Osonoe T, Kikuchi H, Shiba T, Sakaki Y, Hattori M (2001) Genome sequence of an industrial microorganism Streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc Natl Acad Sci U S A 98:12215–12220
Pfeifer B, Hu Z, Licari P, Khosla C (2002) Process and metabolic strategies for improved production of Escherichia coli-derived 6-deoxyerythronolide B. Appl Environ Microbiol 68:3287–3292
Pullan ST, Chandra G, Bibb MJ, Merrick M (2011) Genome-wide analysis of the role of GlnR in Streptomyces venezuelae provides new insights into global nitrogen regulation in actinomycetes. BMC Genomics 12:175
Quaderer R, Omura S, Ikeda H, Cane DE (2006) Pentalenolactone biosynthesis. Molecular cloning and assignment of biochemical function to PtlI, a cytochrome P450 of Streptomyces avermitilis. J Am Chem Soc 128:13036–13037
Seo MJ, Zhu D, Endo S, Ikeda H, Cane DE (2011) Genome mining in Streptomyces. Elucidation of the role of Baeyer-Villiger monooxygenases and non-heme iron-dependent dehydrogenase/oxygenases in the final steps of the biosynthesis of pentalenolactone and neopentalenolactone. Biochemistry 50:1739–1754
Solenberg PJ, Burgett SG (1989) Method for selection of transposable DNA and characterization of a new insertion sequence, IS493, from Streptomyces lividans. J Bacteriol 171:4807–4813
Takamatsu S, Xu LH, Fushinobu S, Shoun H, Komastu M, Cane DE, Ikeda H (2011) Pentalenic acid is a shunt metabolite in the biosynthesis of the pentalenolactone family of metabolites: hydroxylation of 1-deoxypentalenic acid mediated by CYP105D7 (SAV_7469) of Streptomyces avermitilis. J. Antibiot 64:65–71
Takamatsu S, Lin X, Nara A, Komatsu M, Cane DE, Ikeda H (2011) Characterization of a silent sesquiterpenoid biosynthetic pathway in Streptomyces avermitilis controlling epi-isozizaene albaflavenone biosynthesis and isolation of a new oxidized epi-isozizaene metabolite. Microb Biotechnol 4:184–191
Takiguchi Y, Mishima H, Okuda M, Terao M, Aoki A, Fukuda R (1980) Milbemycins, a new family of macrolide antibiotics: fermentation, isolation and physico-chemical properties. J Antibiot 33:1120–1127
Tetzlaff CN, You Z, Cane DE, Takamatsu S, Omura S, Ikeda H (2006) A gene cluster for biosynthesis of the sesquiterpenoid antibiotic pentalenolactone in Streptomyces avermitilis. Biochemistry 45:6179–6186
Uchida M, Takamatsu S, Arima S, Miyamoto KT, Kitani S, Nihira T, Ikeda H, Nagamitsu T (2011) Total synthesis and absolute configuration of avenolide, extracelluar factor in Streptomyces avermitilis. J Antibiot 64:781–787
Ueki M, Suzuki R, Takamatsu S, Takagi H, Uramoto M, Ikeda H, Osada H (2009) Nocardamin production by Streptomyces avermitilis. Actinomycetologica 23:34–39
Wang XJ, Yan YJ, Zhang B, An J, Wang JJ, Tian J, Jiang L, Chen YH, Huang SX, Yin M, Zhang J, Gao AL, Liu CX, Zhu ZX, Xiang WS (2010) Genome sequence of the milbemycin-producing bacterium Streptomyces bingchenggensis. J Bacteriol 192:4526–4527
Xu L, Fushinobu S, Ikeda H, Wakagi T, Shoun H (2009) Crystal structures of cytochrome P450 105P1 from Streptomyces avermitilis: conformational flexibility and histidine-liganded state. J Bacteriol 191:1211–1219
Xu L, Fushinobu S, Takamatsu S, Wakagi T, Ikeda H, Shoun H (2010) Regio- and stereospecificity of filipin hydroxylation sites revealed by crystal structures of cytochrome P450 105P1 and 105D6 from Streptomyces avermitilis. J Biol Chem 285:16844–16853
You Z, Omura S, Ikeda H, Cane DE (2006) Pentalenolactone biosynthesis. Moleclar cloning and assignment of biochemical function to PtlH, a non-heme iron dioxygenase of Streptomyces avermitilis. J Am Chem Soc 128:6566–6567
You Z, Omura S, Ikeda H, Cane DE, Jogl G (2007) Crystal structure of the non-heme iron dioxygenase PtlH in pentalenolactone biosynthesis. J Biol Chem 282:36552–36560
Zhu D, Seo MJ, Ikeda H, Cane DE (2011) Genome mining in Streptomyces. Discovery of an unprecedented P450-catalyzed oxidative rearrangement that is the final step in the biosynthesis of pentalenolactone. J Am Chem Soc 133:2128–2131
Acknowledgments
We thank M. Komatsu, K. Komatsu, H. Koiwai, S. Takamatsu, I. Kozone, M. Izumikawa, and J. Hashimoto for the collaborative experiments. We also thank Y. Shiga for valuable advice about IS elements. We gratefully acknowledge support from a research Grant-in-Aid for Scientific Research on Priority Areas (to H.I.), a research Grant-in-Aid for Scientific Research on Innovative Areas (to H.I.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, a research grant from the Institute for Fermentation, Osaka, Japan (to H.I.), and a research Grant-in-Aid for Scientific Research from the New Energy and Industrial Technology Development Organization (NEDO; to K.S. and H.I.).
Author information
Authors and Affiliations
Corresponding author
Additional information
Dedicated to Prof. Sir David A. Hopwood on the occasion of his 80th birthday and in recognition of his exceptional contributions to Streptomyces genetics and molecular biology, and the honor of his friendship.
Special issue: Genome Mining for Natural Products Discovery.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ikeda, H., Shin-ya, K. & Omura, S. Genome mining of the Streptomyces avermitilis genome and development of genome-minimized hosts for heterologous expression of biosynthetic gene clusters. J Ind Microbiol Biotechnol 41, 233–250 (2014). https://doi.org/10.1007/s10295-013-1327-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-013-1327-x