Abstract
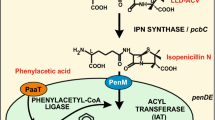
Peroxisomes are eukaryotic organelles surrounded by a single bilayer membrane, containing a variety of proteins depending on the organism; they mainly perform degradation reactions of toxic metabolites (detoxification), catabolism of linear and branched-chain fatty acids, and removal of H2O2 (formed in some oxidative processes) by catalase. Proteins named peroxins are involved in recruiting, transporting, and introducing the peroxisomal matrix proteins into the peroxisomes. The matrix proteins contain the peroxisomal targeting signals PTS1 and/or PTS2 that are recognized by the peroxins Pex5 and Pex7, respectively. Initial evidence indicated that the penicillin biosynthetic enzyme isopenicillin N acyltransferase (IAT) of Penicillium chrysogenum is located inside peroxisomes. There is now solid evidence (based on electron microscopy and/or biochemical data) confirming that IAT and the phenylacetic acid- and fatty acid-activating enzymes are also located in peroxisomes. Similarly, the Acremonium chrysogenum CefD1 and CefD2 proteins that perform the central reactions (activation and epimerization of isopenicillin N) of the cephalosporin pathway are targeted to peroxisomes. Growing evidence supports the conclusion that some enzymes involved in the biosynthesis of mycotoxins (e.g., AK-toxin), and the biosynthesis of signaling molecules in plants (e.g., jasmonic acid or auxins) occur in peroxisomes. The high concentration of substrates (in many cases toxic to the cytoplasm) and enzymes inside the peroxisomes allows efficient synthesis of metabolites with interesting biological or pharmacological activities. This compartmentalization poses additional challenges to the cell due to the need to import the substrates into the peroxisomes and to export the final products; the transporters involved in these processes are still very poorly known. This article focuses on new aspects of the metabolic processes occurring in peroxisomes, namely the degradation and detoxification processes that lead to the biosynthesis and secretion of secondary metabolites.




Similar content being viewed by others
References
Agne B, Meindl NM, Niederhoff K, Einwächter H, Rehling P, Sickmann A, Meyer HE, Girzalsky W, Kunau WH (2003) Pex8p: an intraperoxisomal organizer of the peroxisomal import machinery. Mol Cell 11:635–646
Aharonowitz Y, Cohen G, Martín JF (1992) Penicillin and cephalosporin biosynthetic genes: structure, organization, regulation and evolution. Annu Rev Microbiol 46:461–495
Alvarez E, Cantoral JM, Barredo JL, Díez B, Martín JF (1987) Purification to homogeneity and characterization of the acyl-CoA:6-APA acyltransferase of Penicillium chrysogenum. Antimicrob Agents Chemother 31:1675–1682
Alvarez E, Meesschaert B, Montenegro E, Gutiérrez S, Díez B, Barredo JL, Martín JF (1993) The isopenicillin N acyltransferase of Penicillium chrysogenum has isopenicillin N amidohydrolase, 6-aminopenicillanic acid acyltransferase and penicillin amidase acitivies, all of which are encoded by the single penDE gene. Eur J Biochem 215:323–332
Ashmarina LI, Rusnak N, Miziorko HM, Mitchell GA (1994) 3-Hydroxy-3-methylglutaryl-CoA lyase is present in mouse and human liver peroxisomes. J Biol Chem 269:31929–33132
Baldwin JE, Bird JW, Field RA, O′Callaghan NM, Schoffield CJ (1990) Isolation and partial characterization of ACV synthetase from Cephalosporium acremonium and Streptomyces clavuligerus. J Antibiot 43:1055–1057
Barredo JL, van Solingen P, Díez B, Álvarez E, Cantoral JM, Kattevilder A, Smaal EB, Groenen MA, Veenstra AE, Martín JF (1989) Cloning and characterization of the acyl-coenzyme A: 6-aminopenicillanic-acid-acyltransferase gene of Penicillium chrysogenum. Gene 83:291–300
Barrios-González J, Montenegro E, Martín JF (1993) Penicillin production by mutants resistant to phenylacetic acid. J Ferm Bioeng 76:455–458
Brakhage AA (1997) Molecular regulation of penicillin biosynthesis in Aspergillus (Emericella) nidulans. FEMS Microbiol Lett 148:1–10
Braverman N, Dodt G, Gould SJ, Valle D (1998) An isoform of pex5p, the human PTS1 receptor, is required for the import of PTS2 proteins into peroxisomes. Human Mol Genet 7:1195–1205
Brocard C, Kragler F, Simon MM, Schuster T, Hartig A (1994) The tetratricopeptide repeat-domain of the PAS10 protein of Saccharomyces cerevisiae is essential for binding the peroxisomal targeting signal-SKL. Biochem Biophys Res Commun 204:1016–1022
Cantoral JM, Gutiérrez S, Fierro F, Gil-Espinosa S, van Liempt H, Martín JF (1993) Biochemical characterization and molecular genetics of nine mutants of Penicillium chrysogenum impaired in penicillin biosynthesis. J Biol Chem 268:737–744
Chang CC, Warren DS, Sacksteder KA, Gould SJ (1999) PEX12 binds PEX5 and PEX10 and acts downstream of receptor docking in peroxisomal matrix protein import. J Cell Biol 147:761–774
Dammai V, Subramani S (2001) The human peroxisomal targeting signal receptor, Pex5p, is translocated into the peroxisomal matrix and recycled to the cytosol. Cell 105:187–196
De Duve C, Baudhuin P (1966) Peroxisomes (microbodies and related particles). Physiol Rev 46:323–357
de Hoop MJ, Ab G (1992) Import of proteins into peroxisomes and other microbodies. Biochem J 286:657–669
Delker C, Stenzel I, Hause B, Miersch O, Feussner I, Wasternack C (2006) Jasmonate biosynthesis in Arabidopsis thaliana-enzymes, products, regulation. Plant Biol 8:297–306
Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B (2010) Peroxisomes are signaling platforms for antiviral innate immunity. Cell 141:668–681
Dodt G, Gould SJ (1996) Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J Cell Biol 135:1763–1774
Dotzlaf JE, Yeh WK (1987) Copurification and characterization of deacetoxycephalosporin C synthetase/hydroxylase from Cephalsoporium acremonium. J Bacteriol 169:1611–1618
Dyer JM, McNew JA, Goodman JM (1996) The sorting sequence of the peroxisomal integral membrane protein PMP47 is contained within a short hydrophilic loop. J Cell Biol 133:269–280
Einwächter H, Sowinski S, Kunau WH, Schliebs W (2001) Yarrowia lipolytica Pex20p, Saccharomyces cerevisiae Pex18p/Pex21p and mammalian Pex5pL fulfil a common function in the early steps of the peroxisomal PTS2 import pathway. EMBO Rep 2:1035–1039
Elgersma Y, Kwast L, van den Berg M, Snyder WB, Distel B, Subramani S, Tabak HF (1997) Overexpression of Pex15p, a phosphorylated peroxisomal integral membrane protein required for peroxisome assembly in S.cerevisiae, causes proliferation of the endoplasmic reticulum membrane. EMBO J 16:7326–7341
Epstein E, Nissen SJ, Sutter EG (1991) Indole-3-acetic acid and indole-3-butyric acid in tissues in carrot inoculated with Agrobacterium rhizogenes. J Plant Growth Reg 10:97–1100
Erdmann R, Blobel G (1996) Identification of Pex13p a peroxisomal membrane receptor for the PTS1 recognition factor. J Cell Biol 135:111–121
Erdmann R, Schliebs W (2005) Peroxisomal matrix protein import: the transient pore model. Natl Rev Mol Cell Biol 6:738–742
Evers ME, Trip H, van den Berg MA, Bovenberg RA, Driessen AJ (2004) Compartmentalization and transport in β-lactam antibiotics biosynthesis. Adv Biochem Eng Biotechnol 88:111–135
Fan J, Quan S, Orth T, Awai C, Chory J, Hu J (2005) The Arabidopsis PEX12 gene is required for peroxisome biogenesis and is essential for development. Plant Physiol 139:231–239
Fang Y, Morrell JC, Jones JM, Gould SJ (2004) PEX3 functions as a PEX19 docking factor in the import of class I peroxisomal membrane proteins. J Cell Biol 164:863–875
Ferdinandusse S, Houten SM (2006) Peroxisomes and bile acid biosynthesis. Biochim Biophys Acta 1763:1427–1440
Flaspohler JA, Rickoll WL, Beverley SM, Parsons M (1997) Functional identification of a Leishmania gene related to the peroxin 2 gene reveals common ancestry of glycosomes and peroxisomes. Mol Cell Biol 17:1093–1101
Fransen M, Brees C, Baumgart E, Vanhooren JC, Baes M, Mannaerts GP, Van Veldhoven PP (1995) Identification and characterization of the putative human peroxisomal C-terminal targeting signal import receptor. J Biol Chem 270:7731–7736
Fujiki Y, Matsuzono Y, Matsuzaki T, Fransen M (2006) Import of peroxisomal membrane proteins: the interplay of Pex3p- and Pex19p-mediated interactions. Biochim Biophys Acta 1763:1639–1646
Gabaldón T (2010) Peroxisome diversity and evolution. Philos Trans R Soc Lond B Biol Sci 365:765–773
García-Estrada C, Vaca I, Fierro F, Sjollema K, Veenhuis M, Martín JF (2008) The unprocessed preprotein form IATC103S of the isopenicillin N acyltransferase is transported inside peroxisomes and regulates its self-processing. Fungal Genet Biol 45:1043–1052
García-Estrada C, Vaca I, Ullán RV, van den Berg MA, Bovenberg RA, Martín JF (2009) Molecular characterization of a fungal gene paralogue of the penicillin penDE gene of Penicillium chrysogenum. BMC Microbiol 9:104–119
Geisbrecht BV, Collins CS, Reuber BE, Gould SJ (1998) Disruption of a PEX1-PEX6 interaction is the most common cause of the neurologic disorders Zellweger syndrome, neonatal adrenoleukodystrophy, and infantile Refsum disease. Proc Natl Acad Sci U S A 95:8630–8635
Glover JR, Andrews DW, Rachubinski RA (1994) Saccharomyces cerevisiae peroxisomal thiolase is imported as a dimer. Proc Natl Acad Sci U S A 91:10541–10545
Gould SJ, Kalish JE, Morrell JC, Bjorkman J, Urquhart AJ, Crane DI (1996) Pex13p is an SH3 protein of the peroxisome membrane and a docking factor for the predominantly cytoplasmic PTS1 receptor. J Cell Biol 135:85–95
Grou CP, Carvalho AF, Pinto MP, Wiese S, Piechura H, Meyer HE, Warscheid B, Sá-Miranda C, Azevedo JE (2008) Members of the E2D (UbcH5) family mediate the ubiquitination of the conserved cysteine of Pex5p, the peroxisomal import receptor. J Biol Chem 283:14190–14197
Gutiérrez S, Díez B, Montenegro E, Martín JF (1991) Characterization of the Cephalosporium acremonium pcbAB gene encoding α-aminoadipyl-cysteinyl-valine synthetase, a large multidomain peptide synthetase: linkage to the pcbC gene as a cluster of early cephalosporin biosynthetic genes and evidence of multiple functional domains. J Bacteriol 173:2354–2365
Gutiérrez S, Velasco J, Fernández FJ, Martín JF (1992) The cefG gene of Cephalosporium acremonium is linked to the cefEF gene and encodes a deacetylcephalosporin C acetyltransferase closely related to homoserine O-acetyltransferase. J Bacteriol 174:3056–3064
Gutiérrez S, Fierro F, Casqueiro J, Martín JF (1999) Gene organization and plasticity of the β-lactam genes in different filamentous fungi. Anton van Leeu 75:21–31
Halbach A, Rucktaschel R, Rottensteiner H, Erdmann R (2009) The N-domain of Pex22p can functionally replace the Pex3p N-domain in targeting and peroxisome formation. J Biol Chem 284:3906–3916
Hart DT, Misset O, Edwards SW, Opperdoes FR (1984) Comparison of the glycosomes (microbodies) isolated from Trypanosoma brucei blood stream form and cultured procyclic trypomastigotes. Mol Biochem Parasitol 12:25–35
Hayashi M, Yagi M, Nito K, Kamada T, Nishimura M (2005) Differential contribution of two peroxisomal protein receptors to the maintenance of peroxisomal functions in Arabidopsis. J Biol Chem 280:14829–14835
Heiland I, Erdmann R (2005) Biogenesis of peroxisomes. Topogenesis of the peroxisomal membrane and matrix proteins. FEBS J 272:2362–2372
Hoffmeister D, Keller NP (2007) Natural products of filamentous fungi: enzymes, genes, and their regulation. Nat Prod Rep 24:393–416
Honsho M, Fujiki Y (2001) Topogenesis of peroxisomal membrane protein requires a short, positively charged intervening-loop sequence and flanking hydrophobic segments. J Biol Chem 276:9375–9382
Hu J, Aguirre M, Peto C, Alonso J, Ecker J, Chory J (2002) A role for peroxisomes in photomorphogenesis and development of Arabidopsis. Science 297:405–409
Imazaki A, Tanaka A, Harimoto Y, Yamamoto M, Akimitsu K, Park P, Tsuge T (2010) Contribution of peroxisomes to secondary metabolism and pathogenicity in the fungal plant pathogen. Alternaria Alternata 9:682–694
Islinger M, Li KW, Seitz J, Völkl A, Lüers GH (2009) Hitchhiking of Cu/Zn superoxide dismutase to peroxisomes–evidence for a natural piggyback import mechanism in mammals. Traffic 10:1711–1721
Jami MS, García-Estrada C, Barreiro C, Cuadrado AA, Salehi-Najafabadi Z, Martín JF (2010) The Penicillium chrysogenum extracellular proteome. Conversion from a food-rotting strain to a versatile cell factory for white biotechnology. Mol Cell Proteomics 9:2729–2744
Jedd G, Chua NH (2000) A new self-assembled peroxisomal vesicle required for efficient resealing of the plasma membrane. Nat Cell Biol 2:226–231
Jones JM, Morrell JC, Gould SJ (2004) PEX19 is a predominantly cytosolic chaperone and import receptor for class 1 peroxisomal membrane proteins. J Cell Biol 164:57–67
Kiel JA, Hilbrands RE, van der Klei IJ, Rasmussen SW, Salomons FA, van der Heide M, Faber KN, Cregg JM, Veenhuis M (1999) Hansenula polymorpha Pex1p and Pex6p are peroxisome-associated AAA proteins that functionally and physically interact. Yeast 15:1059–1078
Kiel JA, Hilbrands RE, Bovenberg RAL, Veenhuis M (2000) Isolation of Penicillium chrysogenum PEX1 and PEX6 encoding AAA proteins involved in peroxisome biogenesis. Appl Microbiol Biotechnol 54:234–242
Kiel JA, van den Berg M, Bovenberg RAL, van der Klei IJ, Veenhuis M (2004) Penicillium chrysogenum Pex5p mediates differential sorting of PTS1 proteins to microbodies of the methylotrophic yeast Hansenula polymorpha. Fungal Genet Biol 41:708–720
Kiel JA, van der Klei IJ, van den Berg MA, Bovenberg RAL, Veenhuis M (2005) Overproduction of a single protein, Pc-Pcx11p, results in 2-fold enhanced penicillin production by Penicillium chrysogenum. Fungal Genet Biol 42:154–164
Kiel JA, Veenhuis M, van der Klei IJ (2006) PEX genes in fungal genomes: common, rare or redundant. Traffic 7:1291–1303
Koch A, Schneider G, Lüers GH, Schrader M (2004) Peroxisome elongation and constriction but not fission can occur independently of dynamin-like protein 1. J Cell Sci 117:3995–4006
Koetsier MJ, Jekel PA, van den Berg MA, Bovenberg RA, Janssen DB (2010) Characterization of a phenylacetate-CoA ligase from Penicillium chrysogenum. Biochem J 417:467–476
Kotti TJ, Savolainen K, Helander HM, Yagi A, Novikov DK, Kalkkinen N, Conzelmann E, Hiltunen JK, Schmitz W (2000) In mouse alpha -methylacyl-CoA racemase, the same gene product is simultaneously located in mitochondria and peroxisomes. J Biol Chem 275:20887–20895
Kunze M et al (2006) A central role for the peroxisomal membrane in glyoxylate cycle function. Biochim Biophys Acta 1763:1441–1452
Lamas-Maceiras M, Vaca I, Rodríguez E, Casqueiro J, Martín JF (2006) Amplification and disruption of the phenylacetyl-CoA ligase gene of Penicillium chrysogenum encoding an aryl-capping enzyme that supplies phenylacetic acid to the isopenicillin N acyltransferase. Biochem J 395:147–155
Lazarow PB (2006) The import receptor Pex7p and the PTS2 targeting sequence. Biochim Biophys Acta 1763:1599–1604
Liu F, Ng SK, Lu Y, Low W, Lai J, Jedd G (2008) Making two organelles from one: Woronin body biogenesis by peroxisomal protein sorting. J Cell Biol 180:325–339
Lübbe C, Wolfe S, Demain AL (1986) Isopenicillin N epimerase activity in a high cephalosporin-producing strain of Cephalosporium acremonium. Appl Microbiol Biotechnol 23:367–368
Ma C, Schumann U, Rayapuram N, Subramani S (2009) The peroxisomal matrix import of Pex8p requires only PTS receptors and Pex14p. Mol Biol Cell 20:3680–3689
Managadze D, Würtz C, Sichting M, Niehaus G, Veenhuis M, Rottensteiner H (2007) The peroxin PEX14 of Neurospora crassa is essential for the biogenesis of both glyoxysomes and Woronin bodies. Traffic 8:687–701
Markham P, Collinge AJ (1987) Woronin bodies of filamentous fungi. FEMS Microbiol Rev 46:1–11
Martín JF (2000) Molecular control of expression of penicillin biosynthesis genes in fungi: regulatory proteins interact with a bidirectional promoter region. J Bacteriol 182:2355–2362
Martín JF, Liras P (1989) Enzymes involved in penicillin, cephalosporin and cephamycin biosynthesis. In: Fiechter A (ed) Advances in biochemical engineering/biotechnology. Springer, Berlin, pp 153–187
Martín JF, Ullán RV, Casqueiro FJ (2004) Novel genes involved in Cephalosporin biosynthesis: The three-component isopenicillin N epimerase system. In: Brakhage A (ed) Advances in biochemical engineering-biotechnology. Springer, Berlin, pp 91–109
Martín JF, Ullán RV, García-Estrada C (2010) Regulation and compartmentalization of β-lactam biosynthesis. Microb Biotechnol 3:285–299
Matsumoto N, Tamura S, Fujiki Y (2003) The pathogenic peroxin Pex26p recruits the Pex1p-Pex6p AAA ATPase complexes to peroxisomes. Nat Cell Biol 5:454–460
Matsumura T, Otera H, Fujiki Y (2000) Disruption of the interaction of the longer isoform of Pex5p, Pex5pL, with Pex7p abolishes peroxisome targeting signal type 2 protein import in mammals. Study with a novel Pex5-impaired Chinese hamster ovary cell mutant. J Biol Chem 275:21715–21721
Matsuzono Y, Matsuzaki T, Fujiki Y (2006) Functional domain mapping of peroxin Pex19p: interaction with Pex3p is essential for function and translocation. J Cell Sci 119:3539–3550
Maynard EL, Gatto GJ Jr, Berg JM (2004) Pex5p binding affinities for canonical and noncanonical PTS1 peptides. Proteins 55:856–861
Meinecke M, Cizmowski C, Schliebs W, Krüger V, Beck S, Wagner R, Erdmann R (2010) The peroxisomal importomer constitutes a large and highly dynamic pore. Nat Cell Biol 12:273–277
Montenegro E, Barredo JL, Gutiérrez S, Díez B, Alvarez E, Martín JF (1990) Cloning, characterization of the acyl-CoA:6-amino penicillanic acid acyltransferase gene of Aspergillus nidulans and likage to the isopenicillin N synthase gene. Mol Gen Genet 221:322–330
Motley A, Lumb MJ, Oatey PB, Jennings PR, De Zoysa PA, Wanders RJ, Tabak HF, Danpure CJ (1995) Mammalian alanine/glyoxylate aminotransferase 1 is imported into peroxisomes via the PTS1 translocation pathway. Increased degeneracy and context specificity of the mammalian PTS1 motif and implications for the peroxisome-to-mitochondrion mistargeting of AGT in primary hyperoxaluria type 1. J Cell Biol 131:95–109
Mullen RT, Lee MS, Flynn CR, Trelease RN (1997) Diverse amino acid residues function within the type 1 peroxisomal targeting signal. Implications for the role of accessory residues upstream of the type 1 peroxisomal targeting signal. Plant Physiol 115:881–889
Müller WH, van der Krift TP, Krouwer AJ, Wösten HA, van der Voort LH, Smaal EB, Verkleij AJ (1991) Localization of the pathway of the penicillin biosynthesis in Penicillium chrysogenum. EMBO J 10:489–495
Müller WH, Bovenberg RAL, Groothuis MH, Kattevilder F, Smaal EB, Van der Voort LHM, Verkleij AJ (1992) Involvement of microbodies in penicillin biosynthesis. Biochim et Biophys Acta 1116:210–213
Munck JM, Motley AM, Nuttall JM, Hettema EH (2009) A dual function for Pex3p in peroxisome formation and inheritance. J Cell Biol 187:463–471
Nair DM, Purdue PE, Lazarow PB (2004) Pex7p translocates in and out of peroxisomes in Saccharomyces cerevisiae. J Cell Biol 167:599–604
Neuberger G, Maurer-Stroh S, Eisenhaber B, Hartig A, Eisenhaber F (2003) Motif refinement of the peroxisomal targeting signal 1 and evaluation of taxon-specific differences. J Mol Biol 328:567–579
Nito K, Hayashi M, Nishimura M (2002) Direct interaction and determination of binding domains among peroxisomal import factors in Arabidopsis thaliana. Plant Cell Physiol 43:355–366
Nyathi Y, Baker A (2006) Plant peroxisomes as a source of signalling molecules. Biochim Biophys Acta 1763:1478–1495
Oku M, Sakai Y (2010) Peroxisomes as dynamic organelles: autophagic degradation. FEBS J 277:3289–3294
Opalinski L, Veenhuis M, van der Klei I (2011) Peroxisomes: membrane events accompanying peroxisome proliferation. Int J Biochem Cell Biol 43:847–851
Opalinski L, Kiel JA, Williams C, Veenhuis M, van der Klei IJ (2011) Membrane curvature during peroxisome fission requires Pex11. EMBO J 30:5–16
Opperdoes FR (1984) Localization of the initial steps in alkoxyphospholipid biosynthesis in glycosomes (microbodies) of Trypanosoma Brucei. FEBS Lett 169:35–39
Otera H, Harano T, Honsho M, Ghaedi K, Mukai S, Tanaka A, Kawai A, Shimizu N, Fujiki Y (2000) The mammalian peroxin Pex5pL, the longer isoform of the mobile peroxisome targeting signal (PTS) type 1 transporter, translocates the Pex7p.PTS2 protein complex into peroxisomes via its initial docking site, Pex14p. J Biol Chem 275:21703–21714
Otzen M, Wang D, Lunenborg MG, van der Klei IJ (2005) Hansenula polymorpha Pex20p is an oligomer that binds the peroxisomal targeting signal 2 (PTS2). J Cell Sci 118:3409–3418
Parsons M (2004) Glycosomes: parasites and the divergence of peroxisomal purpose. Mol Microbiol 53:717–724
Perry RJ, Mast FD, Rachubinski RA (2009) Endoplasmic reticulum-associated secretory proteins Sec20p, Sec39p, and Dsl1p are involved in peroxisome biogenesis. Eukaryot Cell 8:830–843
Platta HW, El Magraoui F, Schlee D, Grunau S, Girzalsky W, Erdmann R (2007) Ubiquitination of the peroxisomal import receptor Pex5p is required for its recycling. J Cell Biol 177:197–204
Platta HW, El Magraoui F, Bäumer BE, Schlee D, Girzalsky W, Erdmann R (2009) Pex2 and pex12 function as protein-ubiquitin ligases in peroxisomal protein import. Mol Cell Biol 29:5505–5516
Pracharoenwattana I, Smith SM (2008) When is a peroxisome not a peroxisome? Trends Plant Sci 13:522–525
Praefcke GJ, McMahon HT (2004) The dynamin superfamily: universal membrane tubulation and fission molecules? Natl Rev Mol Cell Biol 5:133–147
Purdue PE, Lazarow PB (1996) Targeting of human catalase to peroxisomes is dependent upon a novel COOH-terminal peroxisomal targeting sequence. J Cell Biol 134:849–862
Purdue PE, Yang X, Lazarow PB (1998) Pex18p and Pex21p, a novel pair of related peroxins essential for peroxisomal targeting by the PTS2 pathway. J Cell Biol 143:1859–1869
Rachubinski RA, Subramani S (1995) How proteins penetrate peroxisomes? Cell 83:525–528
Ramón NM, Bartel B (2010) Interdependence of the peroxisome-targeting receptors in Arabidopsis thaliana: PEX7 facilitates PEX5 accumulation and import of PTS1 cargo into peroxisomes. Mol Biol Cell 21:1263–1271
Ramos FR, López-Nieto MJ, Martín JF (1985) Isopenicillin N synthetase of Penicillium chrysogenum, an enzyme that converts δ-(L-α-aminoadipyl)-L- cysteinyl-d-valine to isopenicillin N. Antimicrob Agents Chemother 27:380–387
Rottensteiner H, Kramer A, Lorenzen S, Stein K, Landgraf C, Volkmer-Engert R, Erdmann R (2004) Peroxisomal membrane proteins contain common Pex19p-binding sites that are an integral part of their targeting signals. Mol Biol Cell 15:3406–3417
Rucktäschel R, Girzalsky W, Erdmann R (2011) Protein import machineries of peroxisomes. Biochim Biophys Acta 1808:892–900
Sakai Y, Oku M, van der Klei IJ, Kiel JA (2006) Pexophagy: autophagic degradation of peroxisomes. Biochim Biophys Acta 1763:1767–1775
Samsom SM, Belagaje R, Blankenship DT, Chapman JL, Perry D, Skatrud PL, van Frank RM, Abraham EP, Baldwin JE, Queener SW, Ingolia TD (1985) Isolation, sequence determination and expression in Escherichia coli of the isopenicillin N synthetase gene from Cephalosporium acremonium. Nature 318:191–194
Samsom SM, Dotzlaf JF, Slisz ML, Becker GW, van Frank RM, Veal LE, Yeh WK, Miller JR, Queener SW, Ingolia TD (1987) Cloning and expression of the fungal expandase/hydroxylase gene involved in cephalosporin biosynthesis. Bio/Technol 5:1207–1214
Saraya R, Veenhuis M, van der Klei IJ (2010) Peroxisomes as dynamic organelles: peroxisome abundance in yeast. FEBS J 277:3279–3288
Scheidegger A, Kuenzi MT, Nuesch J (1984) Partial purification and catalitical properties of a bifunctional enzyme in the biosynthetic pathway of β-lactams in Acremonium chrysogenum. J Antibiot 37:522–531
Schliebs W, Kunau WH (2006) PTS2 co-receptors: diverse proteins with common features. Biochim Biophys Acta 1763:1605–1612
Schneider K, Kienow L, Schmelzer E, Colby T, Bartsch M, Miersch O, Wasternack C, Kombrink E, Stuible H-P (2005) A new type of peroxisomal acyl-coenzyme A synthetase from Arabidopsis thaliana has the catalytic capacity to activate biosynthetic precursors of jasmonic acid. J Bio Chem 280:13962–13972
Schumann U, Wanner G, Veenhuis M, Schmid M, Gietl C (2003) AthPEX10, a nuclear gene essential for peroxisome and storage organelle formation during Arabidopsis embryogenesis. Proc Natl Acad Sci U S A 100:9626–9631
Sichting M, Schell-Steven A, Prokisch H, Erdmann R, Rottensteiner H (2003) Pex7p and Pex20p of Neurospora crassa function together in PTS2-dependent protein import into peroxisomes. Mol Biol Cell 14:810–821
Soukupova M, Sprenger C, Gorgas K, Kunau W-H, Dodt G(1999) Identification and characterization of the human peroxin PEX3. Eur J Cell Biol 78:357–374
Sparkes IA, Brandizzi F, Slocombe SP, El-Shami M, Hawes C, Baker A (2003) An Arabidopsis pex10 null mutant is embryo lethal, implicating peroxisomes in an essential role during plant embryogenesis. Plant Physiol 133:1809–1819
Sparkes IA, Hawes C, Baker A (2005) AtPEX2 and AtPEX10 are targeted to peroxisomes independently of known endoplasmic reticulum trafficking routes. Plant Physiol 139:690–700
Spröte P, Brakhage AA, Hynes MJ (2009) Contribution of peroxisomes to penicillin biosynthesis in Aspergillus nidulans. Eukaryot Cell 8:421–423
Subramani S (1993) Protein import into peroxisomes and biogenesis of the organelle. Annu Rev Cell Biol 9:445–478
Swinkels BW, Gould SJ, Subramani S (1992) Targeting efficiencies of various permutations of the consensus C-terminal tripeptide peroxisomal targeting signal. FEBS Lett 305:133–136
Tabak HF, Murk JL, Braakman I, Geuze HJ (2003) Peroxisomes start their life in the endoplasmic reticulum. Traffic 4:512–518
Tabak HF, van der Zand A, Braakman I (2008) Peroxisomes: minted by the ER. Curr Opin Cell Biol 20:393–400
Teijeira F, Ullán RV, Guerra SM, García-Estrada C, Vaca I, Martín JF (2009) The transporter CefM involved in translocation of biosynthetic intermediates is essential for cephalosporin production. Biochem J 418:113–124
Titorenko VI, Rachubinski RA (1998) Mutants of the yeast Yarrowia lipolytica defective in protein exit from the endoplasmic reticulum are also defective in peroxisome biogenesis. Mol Cell Biol 18:2789–2803
Titorenko VI, Terlecky SR (2011) Peroxisome metabolism and cellular aging. Traffic 12:252–259
Turner JG, Ellis C, Devoto A (2002) The jasmonate signal pathway. Plant Cell 14:153–164
Ullán RV, Liu G, Casqueiro J, Gutiérrez S, Bañuelos O, Martín JF (2002) The cefT gene of Acremonium chrysogenum C10 encodes a putative multidrug efflux pump protein that significantly increases cephalosporin C production. Mol Genet Genom 267:673–683
Ullán RV, Casqueiro J, Bañuelos O, Fernández FJ, Gutiérrez S, Martín JF (2002) A novel epimerization system in fungal secondary metabolism involved in the conversion of isopenicillin N into penicillin N in Acremonium chrysogenum. J Biol Chem 277:46216–46225
Ullán RV, Teijeira F, Guerra SM, Vaca I, Martín JF (2010) Characterization of a novel peroxisome membrane protein essential for conversion of isopenicillin N into cephalosporin C. Biochem J 432:227–236
Van Ael E, Fransen M (2006) Targeting signals in peroxisomal membrane proteins. Biochim Biophys Acta 1763:1629–1638
van den Berg MA, Albang R, Albermann K, Badger JH, Daran JM, Driessen AJ, García-Estrada C, Fedorova ND, Harris DM, Heijne WH, Joardar V, Kiel JA, Kovalchuk A, Martín JF, Nierman WC, Nijland JG, Pronk JT, Roubos JA, van der Klei IJ, van Peij NN, Veenhuis M, von Döhren H, Wagner C, Wortman J, Bovenberg RA (2008) Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nature Biotechnol 26:1161–1168
van de Kamp M, Driessen AJ, Konings WN (1999) Compartmentalization and transport in β-lactam antibiotic biosynthesis by filamentous fungi. Antonie Van Leeuwenhoek 75:41–78
van der Klei IJ, Veenhuis M (1997) Yeast peroxisomes: function and biogenesis of a versatile cell organelle. Trends Microbiol 5:502–509
van der Klei IJ, Veenhuis M (2006) Yeast and filamentous fungi as model organisms in microbody research. Biochim Biophys Acta 1763:1364–1373
van der Lende TR, van de Kamp M, Berg M, Sjollema K, Bovenberg RAL, Veenhuis M et al (2002) Delta-(Lalpha- Aminoadipyl)-L-cysteinyl-d-valine synthetase, that mediates the first committed step in penicillin biosynthesis, is a cytosolic enzyme. Fungal Genet Biol 37:49–55
Velasco J, Gutierrez S, Campoy S, Martin JF (1999) Molecular characterization of the Acremonium chrysogenum cefG gene product: the native deacetylcephalosporin C acetyltransferase is not processed into subunits. Biochem J 337:379–385
Wanders RJ, Waterham HR (2006) Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem 75:295–332
Wang FQ, Liu J, Dai M, Ren ZH, Su CY, He JG (2007) Molecular cloning and functional identification of a novel phenylacetyl-CoA ligase gene from Penicillium chrysogenum. Biochem Biophys Res Commun 360:453–458
Wanner G, Theimer RR (1982) Two types of microbodies in Neurospora crassa. Ann N Y Acad Sci 386:269–284
Watkins PA, Howard AE, Mihalik SJ (1994) Phytanic acid must be activated to phytanoyl-CoA prior to its α-oxidation in rat liver peroxisomes. Biochim Biophys Acta 1214:288–294
Weber H (2002) Fatty acid-derived signals in plants. Trends Plant Sci 7:217–224
Wiemer EAC, Ijlst L, Van Roy J, Wanders RJA, Opperdoes FR (1996) Identification of 2-enoyl coenzyme A hydratase and NADP+- dependent 3-hydroxyacyl-CoA dehydrogenase activity in glycosomes of procyclic Trypanosoma Brucei. Mol Biochem Parasitol 82:107–111
Williams C, van den Berg M, Sprenger RR, Distel B (2007) A conserved cysteine is essential for Pex4p-dependent ubiquitination of the peroxisomal import receptor Pex5p. J Biol Chem 282:22534–22543
Williams C, van den Berg M, Geers E, Distel B (2008) Pex10p functions as an E3 ligase for the Ubc4p-dependent ubiquitination of Pex5p. Biochem Biophys Res Commun 374:620–624
Wolf J, Schliebs W, Erdmann R (2010) Peroxisomes as dynamic organelles: peroxisomal matrix protein import. FEBS J 277:3268–3278
Woodward AW, Bartel B (2005) The Arabidopsis peroxisomal targeting signal type 2 receptor PEX7 is necessary for peroxisome function and dependent on PEX5. Mol Biol Cell 16:573–583
Yu ZL, Liu J, Wang FQ, Dai M, Zhao BH, He JG, Zhang H (2011) Cloning and characterization of a novel CoA-ligase gene from Penicillium chrysogenum. Folia Microbiol (Praha) 56:246–252
Yuan P, Jedd G, Kumaran D, Swaminathan S, Shio H, Hewitt D, Chua NH, Swaminathan K (2003) A HEX-1 crystal lattice required for Woronin body function in Neurospora crassa. Nat Struct Biol 10:264–270
Zolman BK, Yoder A, Bartel B (2000) Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156:1323–1337
Zolman BK, Bartel B (2004) An Arabidopsis indole-3-butyric acid-response mutant defective in PEROXIN6, an apparent ATPase implicated in peroxisomal function. Proc Natl Acad Sci U S A 101:1786–1791
Acknowledgments
This article was supported by grants from the European Union (Eurofungbase LSSG-CT-2005-018964) and EUROFUNGBASE. We thank M. Veenhuis and K. Sjollema (University of Groningen) for the microscopy studies and B. Martín, J. Merino, and A. Mulero for their excellent technical assistance.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Martín, JF., Ullán, R.V. & García-Estrada, C. Role of peroxisomes in the biosynthesis and secretion of β-lactams and other secondary metabolites. J Ind Microbiol Biotechnol 39, 367–382 (2012). https://doi.org/10.1007/s10295-011-1063-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-011-1063-z