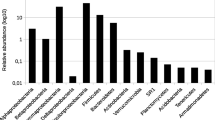
Abstract
We examined the variations of bacterial populations in treated drinking water prior to and after the final chlorine disinfection step at two different surface water treatment plants. For this purpose, the bacterial communities present in treated water were sampled after granular activated carbon (GAC) filtration and chlorine disinfection from two drinking water treatment plants supplying the city of Paris (France). Samples were analyzed after genomic DNA extraction, polymerase chain reaction (PCR) amplification, cloning, and sequencing of a number of 16S ribosomal RNA (rRNA) genes. The 16S rDNA sequences were clustered into operational taxonomic units (OTUs) and the OTU abundance patterns were obtained for each sample. The observed differences suggest that the chlorine disinfection step markedly affects the bacterial community structure and composition present in GAC water. Members of the Alphaproteobacteria and Betaproteobacteria were found to be predominant in the GAC water samples after phylogenetic analyses of the OTUs. Following the chlorine disinfection step, numerous changes were observed, including decreased representation of Proteobacteria phylotypes. Our results indicate that the use of molecular methods to investigate changes in the abundance of certain bacterial groups following chlorine-based disinfection will aid in further understanding the bacterial ecology of drinking water treatment plants (DWTPs), particularly the disinfection step, as it constitutes the final barrier before drinking water distribution to the consumer’s tap.





Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169
Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ (2005) At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl Environ Microbiol 71:7724–7736. doi:10.1128/AEM.71.12.7724-7736.2005
Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL (2008) GenBank. Nucleic Acids Res 36:D25–D30. doi:10.1093/nar/gkn723
Berry D, Xi C, Raskin L (2006) Microbial ecology of drinking water distribution systems. Curr Opin Biotechnol 17:297–302. doi:10.1016/j.copbio.2006.05.007
Brettar I, Höfle MG (2008) Molecular assessment of bacterial pathogens— a contribution to drinking water safety. Curr Opin Biotechnol 19:274–280. doi:10.1016/j.copbio.2008.04.004
Camper AK, LeChevallier MK, Broadaway SC, McFeters GA (1985) Growth and persistence of pathogens on granular activated carbon filters. Appl Environ Microbiol 50:1378–1382
Camper AK, LeChevallier MK, Broadaway SC, McFeters GA (1986) Bacteria associated with granular activated carbon particles in drinking water. Appl Environ Microbiol 52:434–438
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris J, Kulam-Syed-Mohideen AS, McGarrell M, Marsh T, Garrity M, Tiedje JM (2009) The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37:D141–D145. doi:10.1093/nar/gkn879
DeSantis TZ, Hugenholtz P, Keller K, Brodie EL, Larsen L, Picno YM, Phan R, Andersen GL (2006) NAST: a multiple sequence alignment server for comparative analysis of 16S rRNA genes. Nucleic Acids Res 34:W394–W399. doi:10.1093/nar/gkl244
Eichler S, Christen R, Holtje C, Westphal P, Bötel J, Brettar I, Mehling A, Höfle MG (2006) Composition and dynamics of bacterial communities of a drinking water supply system as assessed by RNA- and DNA-based 16S rRNA gene fingerprinting. Appl Environ Microbiol 72:1858–1872. doi:10.1128/AEM.72.3.1858-1872.2006
Falkinham JO, Norton CD, LeChevallier MW (2001) Factors influencing numbers of Mycobacterium avium, Mycobacterium intracellulare and other Mycobacteria in drinking water distribution systems. Appl Environ Microbiol 67:1225–1231. doi:10.1128/AEM.67.3.1225-1231.2001
Felsenstein J (1989) PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164–166
Gerba CP, Nwachuku N, Riley KR (2003) Disinfection resistance of waterborne pathogens on the United States Environmental Protection Agency’s contaminant candidate list (CCL). J Water SRT Aqua 52:81–94
Gomila M, Ramirez A, Gascò J, Lalucat J (2008) Mycobacterium llatzerense sp. nov., a facultatively autotrophic, hydrogen-oxidizing bacterium isolated from haemodialysis water. Int J Syst Evol Microbiol 58:2769–2773. doi:10.1099/ijs.0.65857-0
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Heinemann JA, Rosen H, Savill M, Burgos-Caraballo S, Toranzos GA (2006) Environment arrays: a possible approach for predicting changes in waterborne bacterial disease potential. Environ Sci Technol 40:7150–7156. doi:10.1021/es060331x
Huber T, Faulkner G, Hugenholtz P (2004) Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317–2319. doi:10.1093/bioinformatics/bth226
Humrighouse BW, Santo Domingo JW, Revetta RP, Lamendella R, Kelty CA, Oerther DB (2006) Microbial characterization of drinking water systems receiving groundwater and surface water as the primary sources of water. In: Buchberger SG, Clark RM, Grayman, WM, Uber JG (eds) Water distribution systems analysis symposium 2006. Proceedings of the 8th annual water distribution systems analysis symposium. American Society of Civil Engineering Publication, Reston, Virginia, USA, pp 1–14. doi:10.1061/40941(247)159
Huang J, Wang L, Ren N, Ma F, Ma J (1997) Disinfection effect of chlorine dioxide on bacteria in water. Water Res 31:607–613
Kersters K, De Vos P, Gillis M, Swings J, Vandamme P, Stackebrandt E (2006) Introduction to the Proteobacteria. In: Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E (eds) The prokaryotes: a handbook on the biology of bacteria, vol 5. Springer Science, New York, pp 3–37. doi:10.1007/0-387-30745-1_1
Kistemann T, Claßen T, Koch C, Dangendorf F, Fischeder R, Gebel J, Vacata V, Exner M (2002) Microbial load of drinking water reservoir tributaries during extreme rainfall and runoff. Appl Environ Microbiol 68:2188–2197. doi:10.1128/AEM.68.5.2188-2197.2002
Kormas KA, Neofitou C, Pachiadaki M, Koufostahi E (2009) Changes of the bacterial assemblages throughout an urban drinking water distribution system. Environ Monit Assess. Published online ahead of print on 30 April 2009. doi:10.1007/s10661-009-0924-7
Le Dantec C, Duguet JP, Monteil A, Dumoutier N, Dubrou S, Vincent V (2002) Chlorine disinfection of atypical Mycobacteria isolated from a water distribution system. Appl Environ Microbiol 68:1025–1032. doi:10.1128/AEM.68.3.1025-1032.2002
Le Dantec C, Duguet JP, Montiel A, Dumoutier N, Dubrou S, Vincent V (2002) Occurrence of Mycobacteria in water treatment lines and in water distribution systems. Appl Environ Microbiol 68:5318–5325. doi:10.1128/AEM.68.11.5318-5325.2002
LeChevallier MW, Hassenauer TS, Camper AK, McFeters GA (1984) Disinfection of bacteria attached to granular activated carbon. Appl Environ Microbiol 48:918–923
LeChevallier MW, Welch NJ, Smith DB (1996) Full-scale studies of factors related to coliform regrowth in drinking water. Appl Environ Microbiol 62:2201–2211
Lehtola MJ, Nissinen TK, Miettinen IT, Martikainen PJ, Vartiainen T (2004) Removal of soft deposits from the distribution system improves the drinking water quality. Wat Res 38:601–610. doi:10.1016/j.watres.2003.10.054
Leilei W, Wei C, Tao L (2008) Particle size distribution and property of bacteria attached to carbon fines in drinking water treatment. Water Sci Eng 1:102–111. doi:10.3882/j.issn.1674-2370.2008.02.010
Magic-Knezev A, van der Kooij D (2004) Optimisation and significance of ATP analysis for measuring active biomass in granular activated carbon filters used in water treatment. Water Res 38:3971–3979. doi:10.1016/j.watres.2004.06.017
Magic-Knezev A, Wullings B, Van der Kooij D (2009) Polaromonas and Hydrogenophaga species are the predominant bacteria cultured from granular activated carbon filters in water treatment. J Appl Microbiol 107:1457–1467. doi:10.1111/j.1365-2672.2009.04337.x
Massa S, Armuzzi R, Tosques M, Canganella F, Trovatelli LD (1999) Note: susceptibility to chlorine of Aeromonas hydrophila strains. J Appl Microbiol 86:169–173
Mir J, Morato J, Ribas F (1997) Resistance to chlorine of freshwater bacterial strains. J Appl Microbiol 82:7–18
Norton CD, LeChevallier MW (2000) A pilot study of bacteriological population changes through potable water treatment and distribution. Appl Environ Microbiol 66:268–276
Poitelon J-B, Joyeux M, Welte B, Duguet JP, Prestel E, Lespinet O, DuBow MS (2009) Assessment of phylogenetic diversity of bacterial microflora in drinking water using serial analysis of ribosomal sequence tags. Water Res 43:4197–4206. doi:10.1016/j.watres.2009.07.020
Russel AD (1998) Bacterial resistance to disinfectants: present knowledge and future problems. J Hosp Infect 43:S57–S68
Schloss PD, Handelsman J (2005) Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71:1501–1506. doi:10.1128/AEM.71.3.1501-1506.2005
Schoenen D (2002) Role of disinfection in suppressing the spread of pathogens with drinking water: possibilities and limitations. Water Res 36:3874–3888. doi:10.1016/S0043-1354(02)00076-3
Srinivasan S, Harrington GW, Xagoraraki I, Goel R (2008) Factors affecting bulk to total bacteria ratio in drinking water distribution systems. Water Res 42:3393–3404. doi:10.1016/j.watres.2008.04.025
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849
Stewart MH, Wolfe RL, Means EG (1990) Assessment of the bacteriological activity associated with granular activated carbon treatment of drinking water. Appl Environ Microbiol 56:3822–3829
Szewzyk U, Szewzyk R, Manz W, Schleifer KH (2000) Microbiological safety of drinking water. Annu Rev Microbiol 54:81–127. doi:10.1146/annurev.micro.54.1.81
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599. doi:10.1093/molbev/msm092
Valentin K, John U, Medlin L (2005) Nucleic acid isolation from environmental aqueous samples. Methods Enzymol 395:15–37. doi:10.1016/S0076-6879(05)95002-7
Velten S, Hammes F, Boller M, Egli T (2007) Rapid and direct estimation of active biomass on granular activated carbon through adenosine tri-phosphate (ATP) determination. Water Res 41:1973–1983. doi:10.1016/j.watres.2007.01.021
Whipps CM, Butler WR, Pourahmad F, Watral VG, Kent ML (2007) Molecular systematics support the revival of Mycobacterium salmoniphilum (ex Ross 1960) sp. nov., nom. rev., a species closely related to Mycobacterium chelonae. Int J Syst Evol Microbiol 57:2525–2531. doi:10.1099/ijs.0.64841-0
WHO, OECD (2003) Assessing microbial safety of drinking water: improving approaches and methods. In: Dufour A, Snozzi M, Koster W, Bartram J, Ronchi E (eds). IWA Publishing, London
Wilcox DP, Chang E, Dickson KL, Johansson KR (1983) Microbial growth associated with granular activated carbon in a pilot water treatment facility. Appl Environ Microbiol 46:406–416
Williams MM, Domingo JW, Meckes MC, Kelty CA, Rochon HS (2004) Phylogenetic diversity of drinking water bacteria in a distribution system simulator. J Appl Microbiol 96:954–964. doi:10.1111/j.1365-2672.2004.02229.x
Wilmotte A, Van der Auwera G, De Wachter R (1993) Structure of the 16 S ribosomal RNA of the thermophilic cyanobacterium Chlorogloeopsis HTF (‘Mastigocladus laminosus HTF’) strain PCC7518, and phylogenetic analysis. FEBS Lett 317:96–100. doi:10.1016/0014-5793(93)81499-P
Wolfe RL, Ward NR, Olson BH (1985) Inactivation of heterotrophic bacterial populations in finished drinking water by chlorine and chloramines. Wat Res 19:1393–1403
Zwart G, Crump BC, Kamst-van Agterveld MP, Hagen F, Han SK (2002) Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat Microb Ecol 28:141–155
Acknowledgments
The authors would like to thank Armel Guyonvarch, Erick Denamur, and Eric Delahaye for their comments and suggestions, and also the staff of EAU DE PARIS for their technical assistance. This work was supported by the Association Nationale de la Recherche et de la Technologie de France (ANRT), the municipal water company EAU DE PARIS, the AQUAPHAGE program of the Agence Nationale de la Recherche (ANR, France), and the Centre National de la Recherche Scientifique (CNRS, France).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Poitelon, JB., Joyeux, M., Welté, B. et al. Variations of bacterial 16S rDNA phylotypes prior to and after chlorination for drinking water production from two surface water treatment plants. J Ind Microbiol Biotechnol 37, 117–128 (2010). https://doi.org/10.1007/s10295-009-0653-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-009-0653-5