Abstract
The fungus Rhizopus oryzae converts both glucose and xylose under aerobic conditions into chirally pure l(+)-lactic acid with by-products such as xylitol, glycerol, ethanol, carbon dioxide and fungal biomass. In this paper, we demonstrate that the production of lactic acid by R. oryzae CBS 112.07 only occurs under growing conditions. Deprivation of nutrients such as nitrogen, essential for fungal biomass formation, resulted in a cessation of lactic acid production. Complete xylose utilisation required a significantly lower C/N ratio (61/1) compared to glucose (201/1), caused by higher fungal biomass yields that were obtained with xylose as substrate. Decreasing the oxygen transfer rate resulted in decline of xylose consumption rates, whereas the conversion of glucose by R. oryzae was less affected. Both results were linked to the fact that R. oryzae CBS 112.07 utilises xylose via the two-step reduction/oxidation route. The consequences of these effects for R. oryzae as a potential lactic acid producer are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lactic acid and its derivatives represent an important category of compounds for industries producing food, chemical and pharmaceutical products. Lactic acid can be synthesised chemically, but this results in racemic mixtures of d- and l-isomers. Chirally pure lactic acid can be produced by microbial fermentation and is required for the manufacturing of the biodegradable polymer polylactic acid, which is an alternative to petrochemically derived plastics [6]. Furthermore, the microbial production of lactic acid is of economic importance due to the prospect of using cheap and widely available feedstock materials such as lignocellulose [2].
Lignocellulosic biomass and agricultural residues are rich in polysaccharides such as cellulose (∼35–50%) and hemi-cellulose (∼5–20%), which are embedded in a matrix of lignin [9]. A broad range of physical, chemical and enzymatic treatments can be applied in order to remove/modify the lignin and to hydrolyse the polysaccharides to fermentable monosaccharides [17]. The resulting hydrolysates contain a mixture of hexoses such as glucose and pentoses such as xylose which all form potential substrates for the microbial production of lactic acid.
The Zygomycete fungus Rhizopus oryzae is a well-known producer of chirally pure l(+)-lactic acid from glucose and starch [19, 22, 26, 27] and capable to form cellulolytic and xylanolytic enzymes [3, 18]. In earlier work, we studied the conversion of xylose to lactic acid by ten different wild-type strains of R. oryzae [14]. When cultures of R. oryzae CBS 112.07 were exposed to relatively high xylose concentrations (60–120 g/l), only ±40 g/l xylose was converted to mainly lactic acid. Furthermore, most of the tested R. oryzae xylose-converting cultures showed a lower lactic acid yield in comparison to the cultures grown on glucose as sole carbon source. Carbon dioxide as a product of aerobic respiration was found in higher amounts with the cultures grown on xylose in comparison to the ones on glucose. This indicated that throughout the conversion of xylose by R. oryzae, the respiratory metabolism and xylose-to-lactic acid fermentation are related.
Incomplete substrate utilisation, lactate yields and the requirement of oxygen for product formation are important aspects for the suitability of R. oryzae as lactic acid producer. Therefore, in the present study, the influence of growth and respiration on the conversion of xylose into lactic acid was studied.
Materials and methods
Fungal strain and inoculum preparation
The strains R. oryzae CBS 112.07 (CBS-Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands) and R. oryzae NRRL 395 (NRRL-Northern Regional Research Laboratory, Peoria, IL, USA) were grown on Potato Dextrose Agar at 30°C. Spores were harvested as described previously [14] and stored at −80°C. Mycelial biomass was produced in growth medium containing per litre: glucose 30 g, (NH4)2SO4 1.25 g, KH2PO4 0.6 g, MgSO4·7H2O 0.25 g, ZnSO4·7H2O 0.04 g, Na2HPO4·2H2O 5.37 g and NaH2PO4·H2O 9.64 g. All chemicals, unless indicated otherwise, were purchased from Merck (Darmstadt, Germany). The growth medium was prepared as described previously [14], inoculated with 2 × 105 spores/ml and incubated in a rotary shaker with an agitation rate of 150 rpm at 37°C. Mycelial biomass was harvested by a wash treatment and served as inoculum for shake flask experiments [14].
Shake flask experiments
Composition of the fermentation medium used in the baffled Erlenmeyer shake flask cultures was similar to the growth medium except sodium phosphate salts were replaced by 52 g/l CaCO3 (SA Omya, Benelux, NV, USA) functioning as neutralising agent for lactic acid formation. A filter-sterilised xylose (Sigma-Aldrich, Germany) or glucose solution served as carbon source (cellulose acetate filter with pore size 0.2 μm, Minisart, Sartorius). Fermentation medium was inoculated with washed mycelial biomass [0.1 g mycelial biomass (dry weight) per litre]. Cultures were incubated in a rotary shaker with an agitation rate of 150 rpm and 37°C. Homogenous samples of 1 ml were withdrawn and analysed for substrate and product concentration by High-Pressure Liquid Chromatography.
In order to study the influence of oxygen transfer rate (OTR) on the conversion of glucose and xylose by R. oryzae, reactors with monitoring and control of the oxygen concentration are preferably used. However, as described in previous work [14], growth of the filamentous fungus R. oryzae in submerged controlled stirred fermentor often resulted in heterogeneous growth with mycelium attached around elements in the reactor such as baffles, electrodes and impellers. Therefore, to examine the effect of OTR on the conversion of glucose and xylose by R. oryzae CBS 112.07, another experimental set-up was used where the fungus was cultivated in different medium volumes of 75, 100, 125 and 150 ml.
Preparation of cell-free extract
Fungal cells cultivated in growth medium were centrifuged (5 min at 3,000 rpm) and the wet pellet was stored at −80°C. Prior to use, the pellet was washed with 0.85% NaCl solution to remove residual medium components. The cell-free extract was prepared by freezing the washed cells in liquid nitrogen and grinding in a mortar. An extraction solution containing Bis–Tris buffer pH 6.5, 20 mM MgCl2, 5 mM and ß-mercaptoethanol, 1 mM, was added to the powder (1:1 w/v). After extraction for 1 h, the protein suspension was centrifuged for 15 min at 14,000×g and 4°C to obtain a clear supernatant, which serves as a source of intracellular enzymes.
Enzyme assays
Xylose reductase (XR) and xylitol dehydrogenase (XDH) activity assays were performed in 200-μl volumes. Oxidation of cofactors was monitored in a temperature-controlled spectrophotometer by measuring absorbance at 340 nm. XR activity in cell-free extract (20 μl) was assayed in a sodium phosphate (pH 6.5, 50 mM) buffered mixture containing d-xylose (100 mM) as substrate and NADPH (0.2 mM) as cofactor [25]. The XDH activity in cell-free extract (20 μl) was assayed in a PIPES KOH (pH 7.0, 50 mM) buffered mixture containing d-xylulose (10 mM) as substrate and NADH (0.2 mM) as cofactor [20]. Background activities (assays without xylose or xylulose) were also measured and included into the calculations. One unit of enzyme activity corresponds to 1 nmol of cofactor converted per mg cell-free extract protein per minute.
Xylose isomerase (XI) activity was tested in 1-ml volumes by incubating 20 μl cell-free extract in a 50 mM sodium phosphate buffered (pH 7.0) mixture containing d-xylose (10 mM) and MgCl2 (10 mM) for 17 h at 37°C [8]. The enzymatic activity was stopped by adding 0.05 ml 50% trichloroacetic acid and xylulose was assayed by the cysteine–carbazole–sulphuric acid method [5]. In order to determine the protein content, the proteins in the cell-free extract were separated from the extraction buffer by centrifugation over a membrane (Microcon YM-3, Millipore, USA) and resolved in de-ionised water. The protein content was analysed by the BCA Protein Assay Kit (Pierce, USA) using diluted bovine serum albumin as standard.
Evaluation of gene sequences encoding enzymes involved in xylose metabolism
As part of the Fungal Genome Initiative at the Broad Institute of Harvard and MIT the genome sequence + automated annotation from the R. oryzae strain RA 99-880 has been released on http://www.broad.mit.edu/annotation/fungi/rhizopus_oryzae/. This database enabled us to search for putative genes encoding the enzymes involved in xylose metabolism in R. oryzae. Enzymes of interest are XR (EC 1.1.1.21), XDH (EC 1.1.1.9) and xylulose kinase (XK) (EC 2.7.1.17) catalyzing the conversion from xylose to xylulose-5-P via xylitol and xylulose, respectively, and XI (EC 5.3.1.5), which converts xylose directly in xylulose. A selection of sequences obtained from NCBI and EMBL databases was used to search the R. oryzae genome using the ‘tblastn’ algorithm. For XR, XDH and XK, genes from fungi were selected to create a high probability of finding the corresponding gene. Accession numbers of XR were: Pichia stipitis (X59465), Neurospora crassa (AY876382), Aspergillus niger (AF219625), Candida sp. (AY854501). Accession numbers of XDH were: Pichia stipitis (A16166), Neurospora crassa (XP_325071), Aspergillus oryzae (AB109101), Candida albicans (XP_719434). Accession numbers of XK were: Piromyces sp. (AJ249910), Pichia stipitis (AAF72328), Aspergillus niger (AJ305311), Neurospora crassa (EAA33587), Candida sp. (DQ087275). Since no fungal genes other than the Piromyces sp. gene [8] are known, the search for XI was also performed using genes from different types of organisms having accession numbers: Piromyces sp. (AJ249909), Arabidopsis thaliana (Q9FKK7), Xanthomas campestris (Q8P9T9), Streptomyces albus (P24299), Escherichia coli (P00944), Bacillus subtilis (P04788), Lactobacillus brevis (P29443).
Analytical methods
The treatment of samples and analysis of monosaccharides, organic acids, alcohols and polyols were performed by High-Pressure Liquid Chromatography as described earlier [14]. The cell dry weight determination was performed by filtration of fermentation broth over a pre-weighed filter and drying overnight [14]. The ammonium concentration in fermentation samples was analysed with an Ammonium Test (Spectroquant, Merck, Germany). The chiral purity (%) of lactic acid was determined by derivatisation of all lactates using methanol, after which both enantiomers of methyl lactate were separated on a chiral Gas Chromatography column and detected using a Flame Ionisation Detector. The chiral purity was expressed as the area of the main enantiomer divided by the sum of areas of both enantiomers.
Calculations
The maximal volumetric consumption rate (q smax) is defined as the amount of sugar consumed (g) per litre per hour, whereas the maximal volumetric production rate (q pmax) can be defined as amount of product formed (g) per litre per hour. The yield of lactic acid (Y p/s) is based on the amount of product synthesised (g) divided by the amount of substrate consumed (g). Biomass yield (Y x/s) is calculated by the amount of biomass (g dry weight) divided by the amount of substrate consumed (g). The carbon recovery is calculated with respect to moles of carbon produced divided by moles of carbon consumed as described earlier [14]. Carbon recovery calculations were made to estimate carbon dioxide production by respiration. In these calculations, we included one carbon dioxide for each ethanol produced to compensate for equimolar carbon dioxide formation in ethanol synthesis.
Results
Effect of nutrient limitation on sugar utilisation and product formation by R. oryzae
As described in previous work, cultures of R. oryzae CBS 112.07 grown in fermentation media with relatively high xylose concentrations (60–120 g/l) decreased the consumption rate when approximately 40 g/l was converted, although sufficient carbon source was available [14]. This result suggests that a nutrient, other than xylose, was limiting. According to the composition of the fermentation medium and an assumed fungal biomass composition of CH1.8O0.5N0.16S0.0045P0.0055 [24], it was calculated that nitrogen was the limiting compound.
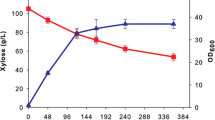
To study the effect of N-limitation on the lactic acid production from xylose by R. oryzae, the fungus was cultivated in fermentation medium containing 80 g/l xylose and 1.25 g/l (NH4)2SO4, corresponding to a C/N ratio of 121/1 (g/g) (Fig. 1a). After 115 h of incubation, approximately 40–45 g/l xylose was converted into 21.1–24.4 g/l lactic acid [chiral purity of 100% l(+)-lactic acid] (Fig. 1b). So, for complete xylose conversion, a C/N ratio of approximately 61/1 is required. The by-products glycerol (3.5–4.0 g/l, Fig. 1c) and xylitol (1.0–1.2 g/l, Fig. 1e) were produced at lower levels. Between 115 and 160 h of incubation, the consumption of xylose by R. oryzae was continued at a low linear rate. To test if this was caused by N-limitation, cultures were pulsed with 2.5 ml (NH4)2SO4 solution, (NH4)2PO4 solution (both 50 g/l) or demi water (blank) after 160 h incubation. In comparison to the blank, the ammonium-pulsed cultures both showed an accelerated xylose consumption rate. Production rates of glycerol (Fig. 1c), ethanol (Fig. 1d) and xylitol (Fig. 1e) increased, whereas no significant lactic acid production (Fig. 1b) was determined after ammonium addition. After 190 h of incubation, the glycerol and ethanol concentrations decreased, suggesting that these compounds can serve as carbon source for R. oryzae and can be used simultaneously with xylose. At the end of the fermentation (240 h), the biomass concentration of the (NH4)2SO4 and (NH4)2PO4-pulsed cultures was 10.2 and 7.6 g/l, respectively, corresponding to a biomass yield (Y x/s) of 0.16 and 0.12 g/g. These biomass concentrations—in the cultures with double ammonium concentration—were approximately twice as high in comparison to the biomass concentration obtained in the culture without the extra ammonium addition.
Former results showed that cessation of product formation, which was observed during growth on xylose and can now be attributed to nitrogen limitation, did not occur under further identical conditions with glucose as substrate [14]. However, utilisation of glucose as carbon source resulted in lower microbial biomass formation compared to xylose, indicating that N-limited conditions might not yet have been established. To investigate this, R. oryzae was cultivated in glucose-containing media with higher C/N ratios. The cultures with C/N ratio of 201/1 and 151/1 converted glucose completely within 70 h of incubation into 74 and 75 g/l lactic acid [chiral purity of 100% l(+)-lactic acid] (Fig. 2a, b), respectively, indicating no N-limitation. The glucose consumption rate of R. oryzae cultivated at higher C/N ratios of 603/1 and 302/1, declined after consumption of 15 and 60 g/l glucose, respectively. By-products such as ethanol (Fig. 2c) and glycerol (Fig. 2d) were formed at different levels depending on the C/N ratio with a maximum of 4.5 and 5.3 g/l, respectively. The addition of extra (NH4)2SO4 restored glucose consumption indicating N-limitation. The concentration of glycerol and ethanol decreased after glucose was completely consumed, suggesting that in the former period catabolite repression by glucose occurred. Figure 2f shows that whereas growth and lactic acid production continued up to 65 h, in all tested R. oryzae cultures the concentration of (NH4)2SO4 was already nearly depleted from the medium after 44 h of incubation. A possible explanation is intracellular accumulation of nitrogen or the formation of reserve material such as glycogen. Furthermore, after 70 h of incubation and prior to the ammonium pulse, the cultures with C/N ratios of 608/1, 302/1, 201/1 and 151/1, showed biomass concentrations of 0.9, 2.2, 3.1 and 3.8 g/l (Fig. 2e), respectively, corresponding to a biomass yield (Y x/s) between 0.03 and 0.06 g/g. To study if growth-associated product formation is a more general phenomenon for R. oryzae, it was also tested for R. oryzae CBS 112.07 with glucose under P-limitation and for R. oryzae NRRL 395 with xylose as substrate under N-limitation. Also, in these cases the conversion of substrate to lactic acid stopped when growth-limiting conditions were reached (results not shown).
Conversion of glucose (a) by R. oryzae CBS 112.07 to lactic acid (b), ethanol (c), glycerol (d) and fungal biomass (e) with initial C/N ratio of 603/1 [0.31 g/l (NH4)2SO4] (dark filled triangle), 302/1 [0.63 g/l (NH4)2SO4] (open square), 201/1 [0.94 g/l (NH4)2SO4] (dark filled diamond) and 151/1 [1.25 g/l (NH4)2SO4] (open triangle). Figure f represents the (NH4)2SO4 concentration in the fermentation medium. The arrow represents the moment of addition of ammonium sulphate
Thus, lactic acid production from both glucose and xylose by R. oryzae CBS 112.07 only occurs under growing conditions. Furthermore, in comparison to glucose, conversion of xylose by R. oryzae required a lower C/N ratio due to a higher biomass yield. This indicates that with xylose as substrate, the energetically more efficient respiration plays a more important role compared to the situation with glucose. The higher respiratory flux could be related to the occurrence of the reduction/oxidation pathway of xylose utilisation in R. oryzae as suggested by subsequent production and consumption of xylitol. In order to better understand the relation between growth, aerobic respiration and lactic acid formation via fermentation, the initial steps in the xylose metabolism of R. oryzae were identified.
Identification of enzymes involved in xylose metabolism by R. oryzae
Most eukaryotic organisms use a two-step reduction/oxidation route to convert xylose via xylitol to xylulose. Some exceptions have been reported where yeast and fungal species employ the isomerase route (XI) [4, 8, 21]. As proposed in Fig. 3, the first enzyme involved in the two-step reduction/oxidation route is the NADPH-dependent XR catalysing the conversion of xylose to xylitol. The polyol xylitol can be excreted or oxidised to xylulose by a NAD-dependent XDH. Further metabolism is from that point onwards identical, starting with the phosphorylation of xylulose to xylulose-5-phosphate by XK and further converted to glyceraldehyde-3-phosphate via the pentose phosphate pathway. The resulting intermediate pyruvate can function as substrate for fermentation via the glycolytic pathway to lactic acid or ethanol, for biomass formation or for oxidation to carbon dioxide and water via respiratory processes [9, 21].
Proposed pathways for the catabolism of xylose, adapted from Schneider (1989) [21]. XI xylose isomerase, XR xylose reductase, XDH xylitol dehydrogenase, XK xylulose kinase. Pyruvate functions as substrate for growth (a), fermentation (b) and/or respiration (c)
To identify the presence of the isomeration route and/or the two-step reduction/oxidation route in R. oryzae CBS 112.07, activities of XI, XR and XDH were assayed by in vitro analysis. Cell-free extracts of R. oryzae CBS 112.07 cultivated in growth medium containing 30 g/l xylose or glucose were used for the enzyme assays. In the glucose culture, R. oryzae converted approximately 30 g/l glucose to 13.5 g/l lactic acid within 48 h of incubation whereas the fungus grown in xylose-containing medium utilises approximately 10 g/l xylose and produces 4.5 g/l lactic acid. The cell-free extract made of fungal biomass cultivated with glucose contained soluble proteins without detectable XI, XR or XDH activity. On the other hand, as shown in Table 1, the cell-free extract of the xylose-converting culture contained detectable XR and XDH activity whereas no XI activity was analysed, indicating the presence of the two-step reduction/oxidation route in R. oryzae.
The rDNA sequence of R. oryzae strain RA 99-880 showed 100% homology with strain CBS 112.07 [1], whereas a lower homology (∼80%) was obtained when comparing with other eukaryotes such as Saccharomyces cerevisiae and Trichoderma. This indicates that the sequence of strain RA 99-880 is representative of strain CBS 112.07. A selection of sequences obtained from NCBI and EMBL databases was used to search the R. oryzae genome using the ‘tblastn’ algorithm. The Blast search on XR (Locus RO3G_16589.1; E-value 3e-100) revealed six possible transcripts (>38% amino acid identity) in the R. oryzae genome all belonging to the aldo/keto reductase family (Pfam domain search). The R. oryzae transcript with the highest homology with the target sequences (47–53% amino acid identity) is likely to be a XDH (Locus RO3G_02257.1; E-value 9e-78). The Blast search on XDH revealed four possible transcripts (>30% amino acid identity) in the R. oryzae genome, all harbouring an alcohol dehydrogenase GroES-like domain and zinc-binding dehydrogenase domain (Pfam domain search). The transcript with the highest homology with the target sequences (44–46% amino acid identity) is likely to be a XDH. The Blast search on XK revealed one possible transcript (>36% amino acid identity) in the R. oryzae genome harbouring the FGGY family of carbohydrate kinases, N-terminal domain and the FGGY family of carbohydrate kinases, C-terminal domain (Pfam domain search). This transcript is likely to be a XK (Locus RO3G_11716.1; E-value 2e-130). The Blast search on XI revealed no possible transcripts in the R. oryzae genome. It seems likely that no gene encoding this enzyme is present in R. oryzae. The findings of this molecular analysis confirm the results obtained by the in vitro enzyme analysis, indicating the presence of the two-step reduction/oxidation route in R. oryzae CBS 112.07.
Effect of oxygen transfer rate on metabolism of R. oryzae
Conversion of xylose via the two-step reduction/oxidation route suggests an enhanced negative effect of oxygen limitation on sugar consumption and product formation compared with glucose [7, 21]. To study the effect of the OTR on the conversion of glucose and xylose by R. oryzae, shake flasks with different medium volumes were used. By altering the volume of the medium in baffled Erlenmeyer flasks, the oxygen transferring liquid/air surface can be varied [10]. In shake flask cultures, we used the carbon recovery calculations in order to estimate the carbon dioxide produced by respiration. Decreasing culture volumes resulted in significantly increased xylose consumption and lactic acid production rates by R. oryzae (Fig. 4a, b). Table 2 shows the maximal xylose consumption rate, q smax of 0.22 to 1.16 g/l/h and the maximal lactic acid production rate, q pmax of 0.16 to 0.78 g/l/h by R. oryzae in different culture volumes. The cultures with volume of 125 and 150 ml resulted, in comparison to the cultures in 75 and 100 ml, in higher concentrations of glycerol and xylitol (Fig. 4c, e). The concentration of these fermentation products decreased at the moment that xylose nearly depleted, indicating that the fungus utilises glycerol and xylitol as carbon source. The lactic acid yields (Y p/s) varied between 0.54 and 0.65 g/g, where the culture of 100 ml yielded the highest amount of lactic acid. The relatively low carbon recoveries show that a significant amount of carbon substrate utilisation can be ascribed to respiratory CO2 production (missing carbon in this calculation).
Effect of different medium volumes (aeration rates) on the conversion of xylose (a) by R. oryzae CBS 112.07 to lactic acid (b), glycerol (c), ethanol (d) and xylitol (e) in a 250-ml baffled shake flasks with medium volumes of 75 (dark filled diamond), 100 (open square), 125 (open triangle) and 150 ml (multiplication sign). Dotted lines represent the start of detectable consumption of xylose
With glucose as carbon source the situation is different. Figure 5a and b show that the conversion rate of glucose into lactic acid by R. oryzae is less affected by the tested culture volume and therefore by the OTR. Lactic acid yields of 0.71–0.79 g/g were calculated and are significantly higher than the yields obtained with xylose (Table 2). By-products such as glycerol (0.3–0.5 g/l) and ethanol (1.2–1.5 g/l) were formed in all cultures up to comparable levels (Fig. 5c, d). When glucose was depleted, the formerly produced glycerol and ethanol concentrations decreased again. Table 2 shows that in comparison to the data obtained with xylose, significantly higher maximal consumption rates (1.77–2.50 g/l/h), lactic acid production rates (1.53–1.97 g/l/h) and lactic acid yields (0.71–0.79 g/g) were obtained with glucose as carbon source. Furthermore, the carbon recovery data were higher in case of glucose cultures (∼95%) compared to xylose cultures (∼75%) suggesting less conversion of substrate to carbon dioxide.
Effect of different medium volumes (aeration rates) on the conversion of glucose (a) by R. oryzae CBS 112.07 to lactic acid (b), glycerol (c) and ethanol (d) in a 250-ml baffled shake flasks with medium volumes of 75 (dark filled triangle), 100 (open square), 125 (open triangle) and 150 ml (multiplication sign). Dotted lines represent the start of detectable consumption of glucose
Discussion
In a previous article we reported that R. oryzae is able to convert xylose into lactic acid and ethanol. Compared to glucose utilisation, the conversion of xylose requires additional enzymatic steps. In this study, we showed several results indicating that R. oryzae utilises xylose via the two-step reduction/oxidation route.
First, in vitro enzyme analysis of cell-free extracts, made from xylose-converting R. oryzae biomass, showed activity of XR and XDH whereas XI activity was lacking. The activity of XR in the cell-free extract was approximately 150-fold higher in comparison to the XDH activity. A possible explanation for this discrepancy in activity can be found in the different hydrophilic characters, determined by the approach described by Kyte and Doolittle (1982) [12], which seems significantly higher for XR than for XDH. As a result, the used extraction procedure may have caused some loss of XDH activity. Furthermore, the stability of the enzymes throughout the extraction procedure was not tested and therefore unknown. Anyhow, detection of XR and XDH indicates the presence of the two-step reduction/oxidation route in R. oryzae.
Second, results from previous study showed that R. oryzae CBS 112.07 secreted xylitol throughout xylose conversion [14]. As soon as the xylose nearly depleted from the medium, the fungus started to utilise the formerly produced xylitol from the medium. Furthermore, R. oryzae cultures converted xylitol, as sole carbon source, to mainly lactic acid (data not shown). As shown in Fig. 3, xylose-fermenting micro-organisms can produce xylitol from xylose either by XR activity or, more unlikely, via activities of XI and XDH. Therefore, the formation and utilisation of xylitol by R. oryzae strongly implies the presence of the two-step reduction/oxidation route to convert xylose.
Finally, comparison of the genome sequence of R. oryzae strain RA 99-880—representative of the used strain CBS 112.07 based on rDNA sequence homology—and sequences of genes representing XR and XDH from the NCBI and EMBL databases showed the presence of genes encoding these enzymes. No homology was found between the genes representing XI and the genome sequence of strain RA 99-880 suggesting once more that the two-step reduction/oxidation route is present in R. oryzae.
The two-step reduction/oxidation route bas been extensively investigated for the production of ethanol by various xylose utilizing yeast species, e.g. Pachysolen tannophilus and Candida shehatae [7, 9, 21]. A consequence of the difference in cofactor dependency of the two key enzymes is that both reactions do not balance out each other’s cofactor requirement. Other metabolic routes have to be applied for this purpose: the pentose phosphate cycle to regenerate NADPH used by XR, and oxygen-requiring respiration to regenerate the NAD used by XDH (Fig. 3). Whereas xylose conversion requires the input of oxygen, ethanol production in these yeasts only occurs under oxygen limitation, with higher ethanol yields at stronger oxygen limitation. Consequently, ethanol production by these yeasts has to occur under suboptimal conditions for both xylose conversion and ethanol production. This rendered the conversion of xylose into ethanol by yeasts harbouring the two-step reduction/oxidation route—both wild type and genetically engineered strains—uneconomical, at least until xylose utilisation by a genetically engineered Saccharomyces cerevisiae was realised and transformed with a gene encoding XI [11].
In our study, a comparable effect of OTR on xylose utilisation and product formation was found for R. oryzae. Decreasing OTR had no significant effect on glucose consumption, but strongly affected the xylose uptake rate. Furthermore, with glucose as substrate ethanol was formed under all experimental conditions whereas with xylose as substrate ethanol production was absent in most cases. So, the disadvantage of the two-step reduction/oxidation pathway for ethanol production from xylose also seems to hold for R. oryzae. This is however not the case for lactic acid since it is produced both under strict aerobic and oxygen-limiting conditions.
Our results showed that the conversion of xylose by R. oryzae strain CBS 112.07 and NRRL 395 not only depends on the availability of a carbon source in the medium but also on the presence of other nutrients, such as nitrogen and phosphorous. This occurred for both xylose and glucose, but at different C/N ratios. These different C/N ratios can be explained by the higher biomass yield on xylose compared to glucose. This, in turn, is caused by the fact that with xylose as substrate, respiration is necessary for the re-oxidation of NADH generated by XDH. Due to the higher energetic efficiency of respiration compared to lactic acid production more fungal biomass can be produced from the same amount of sugar. Or, in other words, less sugar (C) is necessary to convert the same amount of ammonium (N) into biomass, resulting in lower C/N ratios for complete sugar utilisation.
Product formation, limited to conditions at which growth occurs, has—as far as we are aware of—not been reported for a fermentation product as lactic acid. In its natural habitat (mostly soil), R. oryzae will not encounter the high sugar concentrations used in our experiments. Its regulation of sugar metabolism might therefore not be adapted to these situations resulting in lactic acid production as a consequence of overflow metabolism [15]. One could hypothesise that during growth under carbon and energy limitation tight regulation of sugar uptake and/or glycolysis is not necessary because they are rate-limiting anyhow, whereas under non-growing conditions a tight regulation is more important to prevent spillage of resources. This could be an explanation of the growth-associated lactic acid production by R. oryzae. Growth-associated product formation could prove to be a disadvantage for using R. oryzae for lactic acid production since compulsory fungal biomass synthesis has a negative impact on lactic acid yield. The extent of this negative influence depends on the biomass yield on glucose and xylose. In general, the biomass yield for glucose is rather low, 0.03–0.06 g/g, resulting in lactic acid yields of 0.7–0.8 g/g. This corresponds well with lactate yields for homolactic lactobacilli [13]. The biomass yield on xylose is higher due to the relatively higher contribution of respiration to cellular energy generation, resulting in considerably lower lactate yields of 0.55–0.65 g/g. However, considering the fact that lignocellulosic hydrolysates generally contains about two-times higher glucose than xylose concentrations, the negative effect of growth on lactate production from xylose will have a minor impact on overall lactic acid yield.
Current industrial lactic acid production occurs at near neutral pH. At this pH not lactic acid but Ca-lactate is formed. By treatment with sulphuric acid this is converted into lactic acid with CaSO4 (gypsum) as by-product [23]. At the scale necessary for lactic acid as food product it is still possible to deal with gypsum as by-product. However, if lactic acid is to be used at larger scale as building block for the chemical industry it would be advantageous to perform the fermentation process at lower pH, producing lactic acid itself directly. The strains used in contemporary processes are not able to produce lactic acid at these low pH values. Alternatives are sought among yeasts and fungi that are able to grow at lower pH, e.g. R. oryzae [22]. It however appears that, as reviewed by van Maris et al. (2004) [16], cellular export of weak acids such as lactic acid at low pH proceed via active transport and costs metabolic energy (one ATP/acid). This would imply that at low pH lactic acid production does not result in net energy gain and that lactic acid production should always be accompanied by energy-generating processes like alcoholic fermentation or aerobic respiration in order to meet maintenance requirements. So, whereas the isomerase route for xylose conversion is the pathway of choice for synthesis of ethanol because it can function without oxygen input, the situation is not as clear-cut in case of lactic acid production at low pH since this requires oxygen for energy generation anyway.
Rhizopus oryzae is considered to be a potential production organism for lactic acid with advantages compared to lactic acid bacteria, with its ability to convert xylose homolactically, being able to grow without complex nutrients and with a high chiral purity of the produced lactic acid [14]. Because R. oryzae is able to grow at low pH, the possibility to use this micro-organism for the synthesis of lactic acid at low pH has been discussed.
The experiments described in this paper show that lactic acid formation is growth associated. This can be considered as a disadvantage for industrial production; however, it was shown that due to the relatively low biomass yields, this will have only minor consequences for the conversion of lignocellulosic hydrolysates into lactic acid.
Lactic acid production by R. oryzae occurs under aerobic conditions. This can be considered to be a disadvantage, since the productivity of the industrial process may become limited by oxygen transfer. However, if lactic acid production at low pH does not generate net ATP as described by van Maris et al. (2004) [16], lactic acid production has to occur under aerobic conditions anyway.
References
Abe A, Oda Y, Asano K, Sone T (2006) The molecular phylogeny of the genus Rhizopus based on rDNA sequences. Biosci Biotechnol Biochem 70:2387–2393
Åkerberg C, Zacchi G (2000) An economic evaluation of the fermentative production of lactic acid from wheat flour. Bioresour Technol 75:119–126
Bakir U, Yavascaoglu S, Guvenc F, Ersayin A (2001) An endo-β-1,4-xylanase from Rhizopus oryzae: production, partial purification and biochemical characterization. Enzyme Microb Technol 29:328–334
Banerjee S, Archana A, Satyanarayana T (1994) Xylose metabolism in a thermophilic mould Malbranchea pulchella var. sulfurea TMD-8. Curr Microbiol 29:349–352
Dische Z, Borenfreund E (1951) A new spectrophotometric method for the detection and determination of keto sugars and trioses. J Biol Chem 192:583–587
Drumright RE, Gruber PR, Henton DE (2000) Polylactic acid technology. Adv Mater 12:1841–1846
Hahn-Hägerdal, Jeppsson H, Skoog K, Prior BA (1994) Biochemistry and physiology of xylose fermentation by yeast. Enzyme Microb Technol 16:933–943
Harhangi HR, Akhmanova AS, Emmens R, Drift van der C, Laat de WTAM, Dijken van JP, Jetten MSM, Pronk JT, Op den Camp HJM (2003) Xylose metabolism in the anaerobic fungus Piromyces sp. strain E2 follows the bacterial pathway. Arch Microbiol 180:134–141
Jeffries TW (1983) Utilization of xylose by bacteria, yeasts, and fungi. Adv Biochem Eng Biotechnol 27:1–32
Koning de W, Weusthuis RA, Harder W, Dijkhuizen L (1990) Methanol-dependent production of dihydroxyacetone and glycerol by mutants of the methylothropic yeast Hansenula polymorpha blocked in dihydroxyacetone kinase and glycerol kinase. Appl Microbiol Biotechnol 32:693–698
Kuyper M, Winkler AA, Dijken van JP, Pronk JT (2004) Minimal metabolic engineering of Saccharomyces cerevisiae for efficient anaerobic xylose fermentation: a proof of principle. FEMS Yeast Res 4:655–664
Kyte J, Doolittle RF (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157:105–132
Longacre A, Reimers JM, Gannon JE, Wright BE (1997) Flux analysis of glucose metabolism in Rhizopus oryzae for the purpose of increased lactate yields. Fungal Genet Biol 21:30–39
Maas RHW, Bakker RR, Eggink G, Weusthuis RA (2006) Lactic acid production from xylose by the fungus Rhizopus oryzae. Appl Microbiol Biotechnol 72:861–868
Magnuson JK, Lasure LL (2004) Organic acid production by filamentous fungi. In: Lange J, Lange L (eds) Advances in fungal biotechnology for industry, agriculture, and medicine. Kluwer Academic/Plenum Publishers, New York, pp 307–340
Maris van AJA, Konings WN, Dijken van JP, Pronk JT (2004) Microbial export of lactic and 3-hydroxypropanoic acid: implications for industrial fermentation processes. Metab Eng 6:245–255
Mosier N, Wyman C, Dale B, Elander R, Lee YY, Holtzapple M, Ladisch M (2005) Features of promising technologies for pretreatment of lignocellulosic biomass. Bioresour Technol 96:673–686
Murashima K, Nishimura T, Nakamura Y, Koga J, Moriya T, Sumida N, Yaguchi T, Kono T (2002) Purification and characterization of new endo-1,4-β-d-glucanases from Rhizopus oryzae. Enzyme Microb Technol 30:319–326
Ping Huang L, Jin B, Lant P (2005) Direct fermentation of potato starch wastewater to lactic acid by Rhizopus oryzae and Rhizopus arrhizus. Bioprocess Biosyst Eng 27:229–238
Richard P, Toivari MH, Penttilä M (1999) Evidence that the gene YLR070c of Saccharomyces cerevisiae encodes a xylitol dehydrogenase. FEBS Lett 457:135–138
Schneider H (1989) Conversion of pentoses to ethanol by yeast and fungi. Crit Rev Biotechnol 9:1–40
Tay A, Yang ST (2002) Production of L(+)-lactic acid from glucose and starch by immobilized cells of Rhizopus oryzae in a rotating fibrous bed bioreactor. Biotechnol Bioeng 80:1–12
Vaidya AN, Pandey RA, Mudliar S, Suresh Kumar M, Chakrabarti T, Devotta S (2005) Production and recovery of lactic acid for polylactide—an overview. Crit Rev Environ Sci Technol 35:429–467
Van’t Riet K, Tramper J (1991) Basic bioreactor design. Marcel Dekker, New York, Basel, Hong Kong
Witteveen CFB, Busink R, Vondervoort van de P, Dijkema C, Swart K, Visser J (1989) l-Arabinose and d-xylose catabolism in Aspergillus niger. J Gen Microbiol 135:2163–2171
Yin P, Nishina N, Kosakai Y, Yahiro K, Park Y, Okabe M (1997) Enhanced production of l(+)-lactic acid from corn starch in a culture of Rhizopus oryzae using an air-lift bioreactor. J Ferment Bioeng 84:249–253
Zhou Y, Dominguez JM, Cao N, Du J, Tsao GT (1999) Optimization of l-lactic acid production from glucose by Rhizopus oryzae ATCC 52311. Appl Biochem Biotechnol 77–79:401–407
Acknowledgments
This project is supported with a grant of the Dutch Programme EET (Economy, Ecology, Technology) a joint initiative of the Ministries of Economic Affairs, Education, Culture and Sciences and of Housing, Spatial Planning and the Environment. The programme is run by the EET Programme Office, SenterNovem. The authors thank J. Voogt (AFSG, Wageningen, The Netherlands) for his contribution to this work and D. Visser (PURAC, Gorinchem, The Netherlands) for chiral lactic acid purity determination.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Maas, R.H.W., Springer, J., Eggink, G. et al. Xylose metabolism in the fungus Rhizopus oryzae: effect of growth and respiration on l(+)-lactic acid production. J Ind Microbiol Biotechnol 35, 569–578 (2008). https://doi.org/10.1007/s10295-008-0318-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-008-0318-9