Abstract
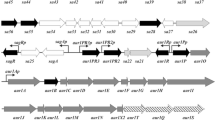
Sphingomonas elodea ATCC 31461 produces gellan, a capsular polysaccharide that is useful as a gelling agent for food and microbiological media. Complementation of nonmucoid S. elodea mutants with a gene library resulted in identification of genes essential for gellan biosynthesis. A cluster of 18 genes spanning 21 kb was isolated. These 18 genes are homologous to genes for synthesis of sphingan polysaccharide S-88 from Sphingomonas sp. ATCC 31554, with predicted amino acid identities varying from 61% to 98%. Both polysaccharides have the same tetrasaccharide repeat unit, comprised of [→4)-α-l-rhamnose-(1→3)-β-d-glucose-(1→4)-β-d-glucuronic acid-(1→4)-β-d-glucose-(1→]. Polysaccharide S-88, however, has mannose or rhamnose in the fourth position and has a rhamnosyl side chain, while gellan has no sugar side chain but is modified by glyceryl and acetyl substituents. Genes for synthesis of the precursor dTDP-l-rhamnose were highly conserved. The least conserved genes in this cluster encode putative glycosyl transferases III and IV and a gene of unknown function, gelF. Three genes (gelI, gelM, and gelN) affected the amount and rheology of gellan produced. Four additional genes present in the S-88 sphingan biosynthetic gene cluster did not have homologs in the gene cluster for gellan biosynthesis. Three of these gene homologs, gelR, gelS, and gelG, were found in an operon unlinked to the main gellan biosynthetic gene cluster. In a third region, a gene possibly involved in positive regulation of gellan biosynthesis was identified.



Similar content being viewed by others
References
Baird JK, Talashek TA, Chang H (1992) Gellan gum: effect of composition on gel properties. In: Phillips GO, Williams PA, Wedlock DJ (eds) Gums and stabilizers for the food industry, vol 6. Oxford University Press, New York, pp 479–487
Chandrasekaran R, Radha A, Thailambal VG (1992) Roles of potassium ions, acetyl and l-glyceryl groups in native gellan double helix: an X-ray study. Carbohydr Res 224:1–17
Chowdhury TA, Lindberg B, Lindquist U, Baird J (1987) Structural studies of an extracellular polysaccharide, S-657, elaborated by Xanthomonas ATCC 53159. Carbohydr Res 164:117–122
Ditta G, Stanfield S, Corbin D, Helinski DR (1980) Broad host range DNA cloning system for Gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci USA 77:7347–7351
Drummelsmith J, Whitfield C (1999) Gene products required for surface expression of the capsular form of the group 1 K antigen in Escherichia coli (O9a:K30). Mol Microbiol 31:1321–1332
Falk C, Jansson P-E, Rinaudo M, Heyraud A, Widmalm G, Hebbar P (1996) Structural studies of the exocellular polysaccharide from Sphingomonas paucimobilis strain I-886. Carbohydr Res 285:69–79
Fialho AM, Monteiro GA, Sa-Correia I (1991) Conjugal transfer of recombinant plasmids into gellan gum-producing and nonproducing variants of Pseudomonas elodea ATCC 31461. Lett Appl Microbiol 12:85–87
Finnie C, Zorreguieta A, Hartley NM, Downie JA (1998) Characterization of Rhizobium leguminosarum exopolysaccharide glycanases that are secreted via a type I exporter and have a novel heptapepide repeat motif. J Bacteriol 180:1691–1699
Gay P, Le Coq D, Steinmetz M, Berkelman T, Kado CI (1985) Positive selection procedure for entrapment of insertion sequence elements in Gram-negative bacteria. J Bacteriol 164:918–921
Guerry P, Van Embden J, Falkow S (1974) Molecular nature of two nonconjugative plasmids carrying drug resistance genes. J Bacteriol 117:619–630
Harding NE, Cleary JM, Cabanas DK, Rosen IG, Kang KS (1987) Genetic and physical analysis of a cluster of genes essential for xanthan gum biosynthesis in Xanthomonas campestris. J Bacteriol 169:2854–2861
Harding NE, Raffo S, Raimondi A, Cleary JM, Ielpi L (1992) Identification, genetic and biochemical analysis of genes involved in synthesis of sugar nucleotide precursors of xanthan gum. J Gen Microbiol 139:447–457
Ielpi L, Couso RO, Dankert MA (1993) Sequential assembly and polymerization of the polyprenol-linked pentasaccharide repeating unit of the xanthan polysaccharide in Xanthomonas campestris. J Bacteriol 175:2490–2500
Jansson PE, Lindberg B, Sandford PA (1983) Structural studies of gellan gum, an extracellular polysaccharide elaborated by Pseudomonas elodea. Carbohydr Res 124:135–139
Jansson PE, Kumar NS, Lindberg B (1986) Structural studies of a polysaccharide (S-88) elaborated by Pseudomonas ATCC 31554. Carbohydr Res 156:165–172
Jay AJ, Colquhoun IJ, Ridout MJ, Brownsey GJ, Morris VJ, Fialho AM, Leitao JH, Sa-Correia I (1998) Analysis of structure and function of gellans with different substitution patterns. Carbohydr Polym 35:179–188
Kamoun S, Tola E, Kamdar H, Kado CI (1992) Rapid generation of directed and unmarked deletions in Xanthomonas. Mol Microbiol 6:809–816
Kang KS, Veeder GT (1982) Polysaccharide S-60 and bacterial fermentation process for its preparation. US Patent 4 326 053
Kang KS, Pettitt DJ (1993) Xanthan, gellan, welan, and rhamsan. In: Whistler RL, BeMiller JN (eds) Industrial gums: polysaccharides and their derivatives, 3rd edn. Academic Press, New York, pp 341–398
Kang KS, Veeder GT, Mirrasoul PJ, Kaneko T, Cottrell IW (1982) Agar-like polysaccharide produced by a Pseudomonas species: production and basic properties. Appl Environ Microbiol 43:1086–1091
Katzen F, Ferreiro DU, Oddo CG, Ielmini MV, Becker A, Puhler A, Ielpi L (1998) Xanthomonas campestris pv. campestris gum mutants: effects on xanthan biosynthesis and plant virulence. J Bacteriol 180:1607–1617
Krontinen VP, Saris P, Sarvas M (1991) A gene (prsA) of Bacillus subtilis involved in a novel, late stage of protein export. Mol Microbiol 5:1273–1283
Kuo MS, Mort AJ, Dell A (1986) Identification and location of l-glycerate, an unusual substituent in gellan gum. Carbohydr Res 56:173–187
Lee EJ, Chandrasekaran R (1991) X-ray and computer modeling studies on gellan-related polymers: molecular structures of welan, S-657, and rhamsan. Carbohydr Res 214:11–24
Lee S-P, Kim T-R, Sinskey AJ (2000) Cloning and sequencing of genes involved in the morphological change of Z. ramigera. J Microbiol Biotechnol 10:161–168
Lenz O, Schwartz E, Dernedde J, Eitinger M, Friedrich B (1994) The Alcaligenes eutrophus H16 hoxX gene participates in hydrogenase regulation. J Bacteriol 176:4385–4393
Liu D, Cole RA, Reeves PR (1996) An O antigen processing function for Wzx (RfbX): a promising candidate for O-unit flippase. J Bacteriol 178:2102–2107
Miller JH (1972) Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
Moorhouse R (1987) Structure/property relationship of a family of microbial polysaccharides. In: Yalpani M (ed) Industrial polysaccharides: genetic engineering, structure/property relations and applications. Elsevier, Amsterdam, pp 187–206
O’Neill MA, Selvendran RR, Morris VJ (1983) Structure of the acidic extracellular gelling polysaccharide produced by Pseudomonas elodea. Carboydr Res 124:123–133
O’Neill MA, Darvill AG, Albersheim P, Chou KJ (1990) Structural analysis of an acidic polysaccharide secreted by Xanthobacter sp. (ATCC 53272). Carbohydr Res 206:289–296
Pollock TJ (1993) Gellan-related polysaccharides and the genus Sphingomonas. J Gen Microbiol 139:1939–1945
Pollock TJ, Armentrout RW (1999) Planktonic/sessile dimorphism of polysaccharide-encapsulated Sphingomonads. J Ind Microbiol Biotechnol 23:436–441
Pollock TJ, Thorne L, Yamazaki M, Mikolajczak M, Armentrout RW (1994) Mechanism of bacitracin resistance in Gram-negative bacteria that synthesize exopolysaccharide. J Bacteriol 176:6229–6237
Pollock TJ, van Workum WAT, Thorne L, Mikolajczak M, Yamazaki M, Kijne JW, Armentrout RW (1998) Assignment of biochemical functions to glycosyl transferase genes which are essential for biosynthesis of exopolysaccharides in Sphingomonas strain S88 and Rhizobium leguminosarum. J Bacteriol 180:586–593
Rahm A, Drummelsmith J, Whitfield C (1999) Conserved organization in the cps gene clusters for expression of Escherichia coli group 1 K antigens: relationship to the colanic acid biosynthesis locus and the cps genes from Klebsiella pneumoniae. J Bacteriol 7:2307–2313
Sa-Correia I, Fialho AM, Videira P, Moreira LM, Marques AR, Albano H (2002) Gellan gum biosynthesis in Sphingomonas paucimobilis ATCC 31461: genes, enzymes and exopolysaccharide production engineering. J Ind Microbiol Biotechnol 29:170–176
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
Skaggs B, Rakitsky W, Phyfferoen A (2000) Method for improved rheological control in cementitious systems. US Patent 6 110 271
Stone KJ, Strominger JL (1971) Mechanism of action of bacitracin: complexation with metal ion and C55-isoprenyl pyrophosphate. Proc Natl Acad Sci USA 68:3223–3227
Staskawicz B, Dahlbeck D, Keen N, Napoli C (1987) Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. Glycinea. J Bacteriol 169:5789–5794
Takeuchi M, Hamana K, Hiraishi A (2001) Proposal of the genus Sphingomonas sensu stricto and three new genera, Sphingobium, Novosphingobium and Sphingopyxis, on the basis of phylogenetic and chemotaxonomic analysis. Int J Syst Evol Microbiol 51:1405–1417
Thorne L, Mikolajczak MJ, Armentrout RW, Pollock TJ (2000) Increasing the yield and viscosity of exopolysaccharides secreted by Sphingomonas by augmentation of chromosomal genes with multiple copies of cloned biosynthetic genes. J Ind Microbiol Biotechnol 25:49–57
Vartak NB, Lin CC, Cleary JM, Fagan MJ, Saier MH Jr (1995) Glucose metabolism in Sphingomonas elodea: pathway engineering via construction of a glucose-6-phosphate dehydrogenase insertion mutant. Microbiology 141:2339–2350
Videira P, Fialho A, Geremia RA, Breton C, Sa-Correia I (2001) Biochemical characterization of the β-1,4-glucuronosyltransferase GelK in the gellan gum-producing strain Sphingomonas paucimobilis ATCC 31461. Biochem J 258:457–464
Vincent C, Doublet P, Grangeasse C, Vaganay E, Cozzone AJ, Duclos B (1999) Cells of Escherichia coli contain a protein-tyrosine kinase, Wzc, and a phosphotyrosine-protein phosphatase, Wzb. J Bacteriol 181:3472–3477
Wugeditsch T, Paiment A, Hocking J, Drummelsmith J, Forrester C, Whitfield C (2001) Phosphorylation of Wzc, a tyrosine autokinase, is essential for assembly of group 1 capsular polysaccharides in Escherichia coli. J Biol Chem 276:2361–2371
Yamazaki M, Thorne L, Mikolajczak M, Armentrout RW, Pollock TJ (1996) Linkage of genes essential for synthesis of a polysaccharide capsule in Sphingomonas strain S88. J Bacteriol 178:2676–2687
Yoshida T, Ayabe Y, Yasunaga M, Usami Y, Habe H, Nojiri H, Omori T (2003) Genes involved in the synthesis of the exopolysaccharide methanolan by the obligate methylotroph Methylobacillus sp strain 12S. Microbiology 149:431–444
Acknowledgements
We thank Joseph M. Cleary for many useful discussions and his careful review of this manuscript. We appreciate the technical assistance of Alan Kanehl, John McQuown and Brian Mueller, and technical discussions with Tom Ramseier and Stan Bower. Sequencing of DNA was accomplished by the Microchemical Core Facility at San Diego State University; Luis Ielpi’s laboratory at the Fundación Instituto Leloir, Buenos Aires, Argentina; Monsanto Company, St. Louis, Mo.; and at Lark Technologies, Houston, Tex.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Harding, N.E., Patel, Y.N. & Coleman, R.J. Organization of genes required for gellan polysaccharide biosynthesis in Sphingomonas elodea ATCC 31461. J IND MICROBIOL BIOTECHNOL 31, 70–82 (2004). https://doi.org/10.1007/s10295-004-0118-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-004-0118-9