Abstract
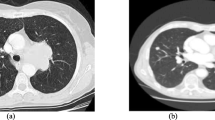
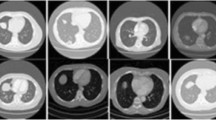
There are lots of work being done to develop computer-assisted diagnosis and detection (CAD) technologies and systems to improve the diagnostic quality for pulmonary nodules. Another way to improve accuracy of diagnosis on new images is to recall or find images with similar features from archived historical images which already have confirmed diagnostic results, and the content-based image retrieval (CBIR) technology has been proposed for this purpose. In this paper, we present a method to find and select texture features of solitary pulmonary nodules (SPNs) detected by computed tomography (CT) and evaluate the performance of support vector machine (SVM)-based classifiers in differentiating benign from malignant SPNs. Seventy-seven biopsy-confirmed CT cases of SPNs were included in this study. A total of 67 features were extracted by a feature extraction procedure, and around 25 features were finally selected after 300 genetic generations. We constructed the SVM-based classifier with the selected features and evaluated the performance of the classifier by comparing the classification results of the SVM-based classifier with six senior radiologists′ observations. The evaluation results not only showed that most of the selected features are characteristics frequently considered by radiologists and used in CAD analyses previously reported in classifying SPNs, but also indicated that some newly found features have important contribution in differentiating benign from malignant SPNs in SVM-based feature space. The results of this research can be used to build the highly efficient feature index of a CBIR system for CT images with pulmonary nodules.






Similar content being viewed by others
References
Matsuki Y, Nakamura K, Watanabe H, Aoki T, Nakata H, Katsuragawa S, Doi K: Usefulness of an artificial neural network for differentiating benign from malignant pulmonary nodules on high-resolution CT: evaluation with receiver operating characteristic analysis. Am J Roentgenol 178(3):657–663, 2002
McNitt-Gray MF, Hart EM, Goldin JG, Yao CW, Aberle DR: A pattern classification approach to characterizing solitary pulmonary nodules imaged on high resolution computed tomography. Proc SPIE 2710:1024–1034, 1996
Nakamura K, Yoshida H, Engelmann R, MacMahon H: Computerized analysis of the likelihood of malignancy in solitary pulmonary nodules with use of artificial neural networks. Radiology 214:823–830, 2000
Shiraishi J, Abe H, Englemann R, Aoyama M: Computer-aided diagnosis to distinguish benign from malignant solitary pulmonary nodules on radiographs: ROC analysis of radiologists′ performance–initial experience. Radiology 227:469–474, 2003
Kawata Y, Niki N, Ohmatsu H, Kusumoto M, et al: Hybrid classification approach of malignant and benign pulmonary nodules based on topological and histogram features. In: Proc MICCAI 297–306, 2000
Silva AC, Paiva AC, Oliveira ACM: Comparison of FLDA, MLP and SVM in diagnosis of lung nodule. Lect Notes Comput Sci 3587:285–294, 2005
Shah SK, McNitt-Gray MF, Rogers SR: Computer aided characterization of the solitary pulmonary nodule using volumetric and contrast enhancement features. Acad Radiol 12(10):1310–1319, 2005
Yamashita K, Matsunobe S, Tsuda T, Nemoto T: Solitary pulmonary nodule: preliminary study of evaluation with incremental dynamic CT. Radiology 194:399–405, 1995
Siegelman SS, Khouri NF, Leo FR: Solitary pulmonary nodules: CT assessment. Radiology 160:307–312, 1986
Müller H, Michoux N, Bandon D: A review of content-based image retrieval system in medical applications-clinical benefits and future directions. Int J Med Informatics 73(1):1–23, 2004
Fisher B, Deserno T, Ott B, et al: Integration of a research CBIR system with RIS and PACS for radiological routine. Proc SPIE 6919:691914–1–691914-10, 2008
Tan Y, Zhang J, Hua Y, Zhang G: Content-based image retrieval in picture archiving and communication system. Proc SPIE 6145:614515–1–614515-8, 2006
Deserno T, Antani S, Long RL: Ontology of gaps in content-based image retrieval. J Digit Imaging (in press), 2007
Depeusinge A, Lavindrasana J, Hidki A, et al: A classification framework for lung tissue categorization. Proc SPIE 6919:69190C1–69190C12, 2008
Silva AC, Carvalho PCP, Gattass M: Diagnosis of lung nodule using semivariogram and geometric measures in computerized tomography images. Comput Methods Programs Biomed 79:31–38, 2005
Haralick RM: Statistical and structural approaches to texture. Proc IEEE 67:786–804, 1979
Clausi DA, Jernigan ME: Designing Gabor filters for optimal texture separability. Pattern Recogn 33:1835–1849, 2000
Manjunath B, Ma W: Texture features for browsing and retrieval of image data. IEEE Trans Pattern Analysis Mach Intell 18(8):837–842, 1996
Unser M: Texture classification and segmentation using wavelet frames. IEEE Trans Image Processing 4:1549–1560, 1995
Kaplan LM, Murenzi R: Texture segmentation using multiscale Hurst features. IEEE Int Conf Image Process 3:205–208, 1997
Joachims T: Text categorization with support vector machines. In: Proceedings of European Conference on Machine Learning (ECML), 1998
Brown M, Grundy W, Lin D, Cristianini N, Sugnet C, Furey T, Ares M, Haussler D: Knowledge-based analysis of microarray gene expression data using support vector machines. 1999. http://www.cse.ucsc.edu/research/compbio/genex/genex.html. Santa Cruz, University of California, Department of Computer Science and Engineering
Shawe-Taylor J, Cristianini N: Kernel methods for pattern analysis, Cambridge: Cambridge University Press, 2004
Fawcett T: ROC graphs: notes and practical considerations for data mining researchers. Technical report HPL-2003-4 HP Labs, 2003.
Canu S, Grandvalet Y, Guigue V, Rakotomamonjy A: SVM and kernel methods Matlab toolbox, Rouen: Perception Systèmes et Information, INSA de Rouen, 2005
Metz CE: ROCKIT software. http://xray.bsd.uchicago.edu/krl/index.htm, 2006
Acknowledgements
The project was supported by the grants from the National Nature Science Foundation of China (grant no. 30570512) and Shanghai Science and Technology Committee (grant no. 064119658, 06SN07111). The authors would like to thank Dr. Xiaojun Ge for providing the CT images used in this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhu, Y., Tan, Y., Hua, Y. et al. Feature Selection and Performance Evaluation of Support Vector Machine (SVM)-Based Classifier for Differentiating Benign and Malignant Pulmonary Nodules by Computed Tomography. J Digit Imaging 23, 51–65 (2010). https://doi.org/10.1007/s10278-009-9185-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10278-009-9185-9