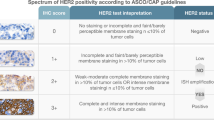
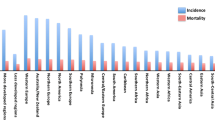
Abstract
Routine clinical and pathological criteria cannot precisely determine the individual prognosis of breast cancer patients, leading to the “over-treatment” of many of them. Three intermediate- or high-throughput, gene-expression-based tools have been developed to make up for this lack of precision. After validation studies on independent series, they are now commercially available. Trials are in progress to assess their real clinical benefits in terms of the determination of prognosis and an enhanced-therapeutic-decision process. These first-generation molecular signatures rely mostly on estrogen receptors, HER2, and proliferation. Would a prognostic algorithm, based on a combination of a well-calibrated determination by immunohistochemistry of these markers, give as much information? This question needs to be addressed.
Résumé
Les critères usuels cliniques et pathologiques ne permettent pas de définir précisément le pronostic individuel des patientes traitées pour cancer du sein et entraînent un « surtraitement » adjuvant d’un grand nombre d’entre elles. Trois outils fondés sur l’expression génique, à moyen ou haut débit, ont été développés pour tenter de pallier cette imprécision. Leurs validations sur des séries indépendantes ont abouti à leur développement commercial. Les études sont en cours pour savoir dans quelle mesure ces signatures génomiques apportent un bénéfice cliniquement perceptible en termes de pronostic et de meilleure indication thérapeutique. Ces signatures moléculaires de première génération reposent essentiellement dans leur composition sur les voies des récepteurs aux estrogènes, du gène HER2 et de la prolifération. Il reste à démontrer qu’une utilisation de marqueurs immunohistochimiques de routine, correctement calibrés et combinés n’apporte pas une information équivalente.
Similar content being viewed by others
Références
Albain KS, Barlow WE, Shak S, et al. (2010) Prognostic and predictive value of the 21-gene Recurrence Score Assay in postmenopausal women with node-positive, oestrogen-receptor-positive breast cancer on chemotherapy: a retrospective analysis of a randomised trial. Lancet Oncol 11(1): 55–65
Buyse M, Loi S, van’t Veer L, et al. (2006) Validation and clinical utility of a 70-gene prognostic signature for women with node-negative breast cancer. J Natl Cancer Inst 98: 1183–1192
Dunkler D, Michiels S, Schemper M (2007) Gene expression profiling: does it add predictive accuracy to clinical characteristics in cancer prognosis? Eur J Cancer 43: 745–751
Eden P, Ritz C, Rose C, et al. (2004) “Good Old” clinical markers have similar power in breast cancer prognosis as microarray gene expression profilers. Eur J Cancer 40: 1837–1841
Ein-Dor L, Kela I, Getz G, et al. (2005) Outcome signature genes in breast cancer: is there a unique set? Bioinformatics 21: 171–178
Ein-Dor L, Zuk O, Domany E (2006) Thousands of samples is needed to generate a robust gene list for predicting outcome in cancer. Proc Natl Acad Sci USA 103: 5923–5928
Elston CW, Ellis IO (1991) Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology 19: 403–410
Esteva FJ, Sahin AA, Cristofanilli M, et al. (2005) Prognostic role of a multigene reverse transcriptase-PCR assay in patients with node-negative breast cancer not receiving adjuvant systemic therapy. Clin Cancer Res 11: 3315–3319
Goldstein LJ, Gray R, Badve S, et al. (2008) Prognostic utility of the 21-gene assay in hormone receptor-positive operable breast cancer compared with classical clinicopathologic features. J Clin Oncol 26: 4063–4071
Habel LA, Shak S, Jacobs MK, et al. (2006) A population-based study of tumor gene expression and risk of breast cancer death among lymph node-negative patients. Breast Cancer Res 8: R25
Hornberger J, Cosler LE, Lyman GH (2005) Economic analysis of targeting chemotherapy using a 21-gene RT-PCR assay in lymph-node-negative, estrogen-receptor-positive, early-stage breast cancer. Am J Manag Care 11: 313–324
Lander ES, Linton LM, Birren B, et al. (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921
Loi S, Haibe-Kains B, Desmedt C, et al. (2007) Definition of clinically distinct molecular subtypes in estrogen receptorpositive breast carcinomas through genomic grade. J Clin Oncol 25: 1239–1246
Lyman GH, Cosler LE, Kuderer NM, Hornberger J (2007) Impact of a 21-gene RT-PCR assay on treatment decisions in early-stage breast cancer: an economic analysis based on prognostic and predictive validation studies. Cancer 109: 1011–1018
Mamounas E, Budd GT, Miller K (2008) Incorporating the Oncotype DX breast cancer assay into community practice: an expert Q & A and case study sampling. Clin Adv Hematol Oncol 6: s1–s8
Mook S, Schmidt MK, Viale G, et al. (2008) The 70-gene prognosis-signature predicts disease outcome in breast cancer patients with 1–3 positive lymph nodes in an independent validation study. Breast Cancer Res Treat
Mook S, Schmidt MK, Weigelt B, et al. (2009) The 70-gene prognosis signature predicts early metastasis in breast cancer patients between 55 and 70 years of age. Ann Oncol [Epub ahead of print]
Paik S, Shak S, Tang G, et al. (2004) A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 351: 2817–2826
Paik S, Tang G, Shak S, et al. (2006) Gene expression and benefit of chemotherapy in women with node-negative, estrogen receptor-positive breast cancer. J Clin Oncol 24: 3726–3734
Ravdin PM (1995) A computer based program to assist in adjuvant therapy decisions for individual breast cancer patients. Bull Cancer 82(Suppl 5): 561s–564s
Reyal F, van Vliet MH, Armstrong NJ, et al. (2008) A comprehensive analysis of prognostic signatures reveals the high predictive capacity of the proliferation, immune response and RNA splicing modules in breast cancer. Breast Cancer Res 10: R93
Schena M, Heller RA, Theriault TP, et al. (1998) Microarrays: biotechnology’s discovery platform for functional genomics. Trends Biotechnol 16: 301–306
Schena M, Shalon D, Davis RW, Brown PO (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270: 467–470
Sorlie T, Perou CM, Tibshirani R, et al. (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA 98: 10869–10874
Sotiriou C, Wirapati P, Loi S, et al. (2006) Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst 98: 262–272
van de Vijver MJ, He YD, van’t Veer LJ, et al. (2002) A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 347: 1999–2009
van’t Veer LJ, Dai H, van de Vijver MJ, et al. (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415: 530–536
Venter JC, Adams MD, Myers EW, et al. (2001) The sequence of the human genome. Science 291: 1304–1351
Wirapati P, Sotiriou C, Kunkel S, et al. (2008) Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res 10: R65
Wittner BS, Sgroi DC, Ryan PD, et al. (2008) Analysis of the MammaPrint breast cancer assay in a predominantly postmenopausal cohort. Clin Cancer Res 14: 2988–2993
Wolf I, Ben-Baruch N, Shapira-Frommer R, et al. (2008) Association between standard clinical and pathologic characteristics and the 21-gene recurrence score in breast cancer patients: a population-based study. Cancer 112: 731–736
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Reyal, F., Pierga, J.Y., Salmon, R.J. et al. Le point sur les signatures moléculaires dans le cancer du sein. Oncologie 12, 263–268 (2010). https://doi.org/10.1007/s10269-010-1876-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10269-010-1876-9