Abstract
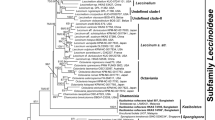
Balanophora japonica and B. yakushimensis are two putatively agamospermic taxa previously reported from southern Japan. Their inflorescences superficially represent those of B. laxiflora and B. fungosa. In this study we confirmed their presence in Taiwan by morphological and phylogenetic analysis using nuclear 18S rDNA and nrITS sequences with related taxa. B. japonica, B. yakushimensis, and B. laxiflora formed a well-supported clade that is distinct from other Balanophora. All three taxa also show considerable differences on morphological and nucleotide sequence differences, therefore the name of B. yakushimensis is retained. The results provide new insights on the intrageneric classification of Balanophora and suggest the positioning of female flowers should be down-weighted. We also successfully identify the hosts of B. japonica and B. yakushimensis by amplifying chloroplast matK sequences from the connected root tissues. The results showed that B. japonica parasitizes on Symplocos species, and that B. yakushimensis parasitizes on Distylium racemosum in Japan and Schima superba in Taiwan’s population.


Similar content being viewed by others
References
Barbrook AC, Howe CJ, Purton S (2006) Why are plastid genomes retained in non-photosynthetic organisms? Trends Plant Sci 11:101–108
Brunner I, Brodbeck S, Buchler U, Sperisen C (2001) Molecular identification of fine roots of trees from the Alps: reliable and fast DNA extraction and PCR-RFLP analyses of plastid DNA. Mol Ecol 10:2079–2087
de Queiroz A, Gatesy J (2007) The supermatrix approach to systematics. Trends Ecol Evol 22:34–41
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Eberwein R, Nickrent DL, Weber A (2009) Development and morphology of flowers and inflorescences in Balanophora papuana and B. elongata (Balanophoraceae). Am J Bot 96:1055–1067
Fagerlind F (1948) Bau und entwicklung der vegetativen organe von Balanophora. Kungl Svenska Vetenskapsakademiens Handlingar 25:1–72
Frank DA, Pontes AW, Maine EM, Caruana J, Raina R, Raina S, Fridley JD (2010) Grassland root communities: species distributions and how they are linked to aboveground abundance. Ecology 91:3201–3209
Hansen B (1972) The genus Balanophora J. R. & G. Forster a taxonomic monograph. Dansk Botanisk Arkiv 28:1–188
Hansen B (1982) The Balanophoraceae of the Pacific. Acta Phytotaxon Geobot 18:92–102
Hansen B (1999) Balanophora species published 1971–1998, mostly from China and Japan. Nord J Bot 19:641–642
Hatushima S (1971) A new noteworthy species of Balanophora from Kyushu. J Geobot 19:60–62
Horikawa Y (1972) Atlas of the Japanese flora—an introduction to plant sociology of East Asia. Gakken Co., Tokyo
Horikawa Y (1976) Atlas of the Japanese flora II—an introduction to plant sociology of East Asia. Gakken Co., Tokyo
Huang S, Murata J (2003) Balanophoraceae. Flora of China, vol 5. Missouri Botanical Garden, pp 272–276
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Kuwada Y (1928) An occurrence of restitution nuclei in the formation of embryo sacs in Balanophora japonica. Bot Mag Tokyo 42:117–129
Linder CR, Moore LA, Jackson RB (2000) A universal molecular method for identifying underground plant parts to species. Mol Ecol 9:1549–1559
Maddison DR, Maddison WP (2000) MacClade 4: analysis of phylogeny and character evolution. Sinauer Associates, Sunderland
Minamitani T (2009) Kyushu plant notes (9): the appearance of Balanophora yakushimensis on the mainland of Kyushu. J Miyazaki Plant Res 11:7–10
Murata J (1988) Morphology and distribution of Balanophora fungosa J. R. et G. Forst. (Balanophoraceae). J Jpn Bot 63:201–210
Murata J (1990) Agamic species of Balanophora in Japan. Mem Natl Sci Mus Tokyo 23:43–50
Murata J (1992) Female flowers of Balanophora kiusiana. J Jpn Bot 67:166–168
Nickrent DL, Duff RJ (1996) Molecular studies of parasitic plants using ribosomal RNA. In: Moreno MT, Cubero JI, Berner D, Joel D, Musselman LJ, Parker C (eds) Advances in parasitic research. Junta de Andalucia, Dirección General de Investigación Agraria, Cordoba, pp 28–52
Nickrent DL, Garcia MA (2009) On the brink of holoparasitism: plastome evolution in dwarf mistletoes (Arceuthobium, Viscaceae). J Mol Evol 68:603–615
Nickrent DL, Starr EM (1994) High-rates of nucleotide substitution in nuclear small subunit (18S) rDNA from holoparasitic flowering plants. J Mol Evol 39:62–70
Nickrent DL, Ouyang Y, Duff RJ, de Pamphilis CW (1997) Do nonasterid holoparasitic flowering plants have plastid genomes? Plant Mol Biol 34:717–729
Nickrent DL, Blarer A, Qiu YL, Yidal-Russell R, Anderson FE (2004) Phylogenetic inference in Rafflesiales: the influence of rate heterogeneity and horizontal gene transfer. BMC Evol Biol 4:40
Nickrent DL, Der JP, Anderson FE (2005) Discovery of the photosynthetic relatives of the “Maltese mushroom” Cynomorium. BMC Evol Biol 5:28
Posada D, Buckley TR (2004) Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol 53:793–808
Sunaryo (1992) Morphologisch-anatomische Untersuchungen an den Knollen von Balanophora fungosa J. R. & G. Forst. ssp. indica (Arn.) B. Hansen var. globosa (Jungh.) B. Hansen und Balanophora elongata Bl. Fachbereichs Biologie. Ph.D. Philipps-Universität Marburg, Marburg, p 122
Swofford DL (2002) PAUP*. Phylogenetic analysis using parsimony (*and other methods). Sinauer Associates, Sunderland
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL-X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Watanabe K (1942) Morphologisch-biologische studien uber Balanophoraceen in Nippon ausgenommen Taiwan (I). J Jpn Bot 18:250–260
Watanabe K, Akuzawa E (1982) Balanophora. In: Satake Y (ed) Wild flowers of Japan, herbaceous plants, vol 2. Heibonsha, Tokyo, pp 12–13
Yahara T, Ohba H, Murata J, Iwatsuki K (1986) Taxonomic review of vascular plants endemic to Yakushima Island, Japan. J Fac Sci Univ Tokyo III 14:82–83
Acknowledgments
We deeply appreciate Dr. Shu-Chuan Hsiao, Mr. Tadashi Minamitani and Jiunn-Yih Huang for their help in specimen collections. We also thank the technical assistance of Technology Commons, College of Life Science, NTU with SEM, and also the staffs at the herbaria of Harvard University (A, GH) and Biodiversity Research Center, Academia Sinica (HAST) for the help on literature survey and providing specimen samples. We thank David W. Taylor for helps on editing the manuscript. This study was partly supported by grants from National Science Council, Taiwan (NSC 96-2621-B-002-008-MY3 and 99-2918-I-002-020).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10265_2011_447_MOESM3_ESM.tif
S3 Unrooted maximum likelihood tree of Balanophora species, including B. reflexa (EU598798), using nr18S/nrITS regions, but excluding non-alignable nrITS1 region (TIFF 789 kb)
10265_2011_447_MOESM4_ESM.tiff
S4 Bracts of B. japonica and B. yakushimensis and SEM photos of epidermal cells on the margin of bract apex. a, bracts of B. japonica, showing smooth margin; b, cells in B. japonica are uniform in morphology with equivalent of length and width; c, bracts of B. yakushimensis, showing irregular crenate margins; d, cells on the margin of B. yakushimensis are 1.5-2 times longer than wide (TIFF 3169 kb)
Rights and permissions
About this article
Cite this article
Su, HJ., Murata, J. & Hu, JM. Morphology and phylogenetics of two holoparasitic plants, Balanophora japonica and Balanophora yakushimensis (Balanophoraceae), and their hosts in Taiwan and Japan. J Plant Res 125, 317–326 (2012). https://doi.org/10.1007/s10265-011-0447-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-011-0447-5