Abstract
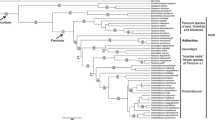
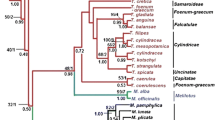
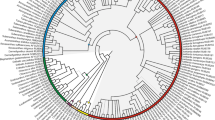
A phylogenetic analysis of Violaceae is presented using sequences from rbcL, atpB, matK and 18S rDNA from 39 species and 19 genera. The combined analysis of four molecular markers resulted in only one most parsimonious tree, and 33 of all 38 nodes within Violaceae are supported by a bootstrap proportion of more than 50%. Fusispermum is in a basal-most position and Rinorea, Decorsella, Rinoreocarpus and the other Violaceae are successively diverged. The monogeneric subfamily Fusispermoideae is supported, and it shares a number of plesiomorphies with Passifloraceae (a convolute petal aestivation, actinomorphic flowers and connate filaments). The other monogeneric subfamily Leonioideae is sunken within the subfamily Violoideae and is sister to Gloeospermum, sharing some seed morphological characteristics. The present molecular phylogenetic analysis suggests that the convolute, apotact and quincuncial petal aestivation is successively derived within the family. The evolutionary trends of the other morphological characteristics, such as a filament connation, the number of carpels and floral symmetry, are discussed.


Similar content being viewed by others
References
APG II (2003) An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG II. Bot J Linn Soc 141:399–436
Arbo MM (2007) Turneraceae. In: Kubitzki K (ed) The families and genera of vascular plants. Springer, Berlin
Chase MW, Soltis DE, Olmstead RG, Morgan D, Les DH, Mishler BD, Duvall MR, Price RA, Hills HG, Qiu Y-L, Kron KA, Pettig JH, Conti E, Palmer JD, Manhart JR, Sytsma KJ, Michaels HJ, Kress WJ, Karol KG, Clark WD, Hedrén M, Gaut BS, Jansen RK, Kim K-J, Wimpee CF, Smith JF, Furnier GR, Strauss SH, Xiang QY, Plunkett GM, Soltis PS, Swensen SM, Williams SE, Gadek PA, Quinn CJ, Eguiarte LE, Golenberg E, Learn GH Jr, Graham SW, Barrett SCH, Dayanandan S, Albert VA (1993) Phylogenetics of seed plants: an analysis of nucleotide sequences from the plastid gene rbcL. Ann Missouri Bot Gard 80:528–580
Chase MW, Zmarzty S, Lledó MD, Wurdack KJ, Swensen SM, Fay MF (2002) When in doubt, put it in Flacourtiaceae: a molecular phylogenetic analysis based on plastid rbcL DNA sequences. Kew Bull 57:141–181
Cronquist A (1981) An integrated system of classification of flowering plants. Columbia Univ Press, New York
Davis CC, Webb CO, Wurdack KJ, Jaramillo CA, Donoghue MJ (2005) Explosive radiation of Malpighiales supports a Mid-Cretaceous origin of modern tropical rain forests. Amer Naturalist 165:E36–E65
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Feng M, Ballard HE (2005) Molecular systematic, floral developmental and anatomical revelations on generic relationships and evolutionary patterns in the Violaceae. XVII International Botanical Congress (Abstract) pp 169
Feuillet C, MacDougal JM (2007) Passifloraceae. In: Kubitzki K (ed) The families and genera of vascular plants. Springer, Berlin
Grey-Wilson CB (1986) Flora of tropical East Africa, Violaceae. Published on behalf of the East African governments by Balkema, Rotterdam
Hekking WHA (1984) Studies on neotropical Violaceae: the genus Fusispermum. Proc Kon Nederl Akad Wetensch Bot C 87:121–130
Hekking WHA (1988) Flora neotropica monograph 46, Violaceae Part I–Rinorea and Rinoreocarpus. New York Botanical Garden, New York
Hodges SA, Ballard HE, Arnold ML, Chase MW (1995) Generic relationships in the Violaceae: data from morphology, anatomy, chromosome numbers and rbcL sequences. Amer J Bot Suppl 82:136
Huelsembeck JP, Ronquist F (2001) MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755
Kubitzki K (2007) Malesherbiaceae. In: Kubitzki K (ed) The families and genera of vascular plants. Springer, Berlin
Maddison DR, Maddison WP (2003) MacClade 4.08 for MacOS X. Analysis of phylogeny and character evolution. Sinauer, Sunderland, Massachusetts, USA
Mason-Gamer RJ, Kellogg EA (1996) Testing for phylogenetic conflict among molecular data sets in the tribe Triticeae (Gramineae). Syst Biol 45(4):524–545
Melchior H (1925) Violaceae. In Die natürlichen Pflanzenfamilien (ed) 2. vol 21. (eds). A Engler and K Prantl. Wilhelm Engelmann Leipzig, Germany
Munzinger JK, Ballard HE (2003) Hekkingia Violaceae, a new arborescent violet genus from French Guiana, with a key to genera in the family. Syst Bot 28(2):345–351
Nylander JAA (2004) MrModeltest, version 2. Computer program distributed by the author. Evolutionary Biology Centre, Uppsala University, Uppsala
Ronquist F, Huelsembeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Savolainen V, Fay MF, Albach DC, Backlund A, van der Bank M, Cameron KM, Johnson SA, Lledó MD, Pintaud JC, Powell M, Sheahan MC, Soltis DE, Soltis PS, Weston P, Whitten WM, Wurdack KJ, Chase MW (2000a) Phylogeny of the eudicots: a nearly complete familial analysis based on rbcL gene sequences. Kew Bull 55:257–309
Savolainen V, Chase MW, Hoot SB, Morton CM, Soltis DE, Bayer C, Fay MF, Bruijn AY de, Sullivan S, Qiu YL, (2000b) Phylogenetics of flowering plants based on combined analysis of plastid atpB and rbcL gene sequences. Syst Biol 49:306–362
Soltis DE, Soltis PS, Chase MW, Mort ME, Albach DC, Zanis M, Savolainen V, Hahn WH, Hoot SB, Fay MF, Axtell M, Swensen SM, Prince LM, Kress WJ, Nixon KC, Farris JS (2000) Angiosperm phylogeny inferred from 18S rDNA, rbcL, and atpB sequences. Bot J Linn Soc 133:381–461
Soltis DE, Gitzendanner MA, Soltis PS (2007) A 567–taxon data set for angiosperms: the challenges posed by Bayesian analyses of large data sets. Int J Plant Sci 168(2):137–157
Swofford DL (2001) PAUP: phylogenetic analysis using parsimony, ver 4.0b10. Sinauer, Sunderland, Massachusetts, USA
Takhtajan A (1997) Diversity and classification of flowering plants. Columbia Univ Press, New York
Taton A (1969) Violaceae, Flore du Congo du Rwanda et du Burundi, Jardin botanique national de Belgique, Bruxelles
Tokuoka T, Tobe H (2006) Phylogenetic analyses of Malpighiales using plastid and nuclear DNA sequences, with particular reference to the embryology of Euphorbiaceae sensu stricto. J Plant Res 119:599–616
Tokuoka T (2007) Molecular phylogenetic analysis of Euphorbiaceae sensu stricto based on plastid and nuclear DNA sequences and ovule and seed character evolution. J Plant Res 120:511–522
Acknowledgments
I am grateful to the curators of MO, L and KYO, for their assistance in obtaining material used in this study, and to Hiroshi Tobe (KYO) and Louis P. Ronse De Craene (E) for helpful comments on earlier drafts of the manuscript. The study was supported by a grant-in-aid for scientific research from the Japan Society for the Promotion of Science (17770069).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Tokuoka, T. Molecular phylogenetic analysis of Violaceae (Malpighiales) based on plastid and nuclear DNA sequences. J Plant Res 121, 253–260 (2008). https://doi.org/10.1007/s10265-008-0153-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10265-008-0153-0