Abstract
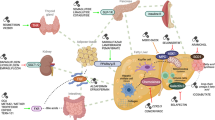
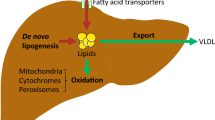
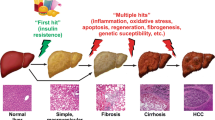
Longitudinal studies have improved current diagnostics and management of metabolic associated fatty liver disease (MAFLD) patients by liver biopsy and therapeutic intervention, yet the deficiency of biomarker spectrum for dissecting subtypes largely hinders the symptomatic treatment. We originally enriched serum from peripheral blood of 618 healthy donors (HD) and 580 MAFLD (400 NAFL, 180 NASH) patients according to multiple clinicopathological indicators. Microarray profiling and qRT-PCR were conducted to identify lncRNAs as candidate biomarkers of MAFLD. Then, we analyzed the matching score of the indicated lncRNA with CAP or MAFLD-associated pathological parameters as well. Additionally, we took advantage of interaction network together with gene expression profiling analysis to further explore the underlying target genes of the identified lncRNA. Herein, we found CAP in nearly all of the NAFL (399/400) and NASH (179/180) patients was higher than that in the HDs (611/618). The differentially expressed lncRNAs were involved in multiple metabolic or immunologic processes by regulating MAFLD-associated pathways. Of them, serum lncPRYP4-3 was identified as a novel candidate biomarker of MAFLD, which was further confirmed by correlation analysis with clinical indicators. Thereafter, we deduced PRS4Y2 was a candidate target of lncPRYP4-3 and mediated the dysfunction in NAFL and NASH patients. Serum lncPRYP4-3 served as a novel biomarker of MAFLD and helped distinguish the subtypes and benefit precise intervention therapy. Our findings also provided overwhelming new evidence for the alteration in biological processes and gene ontology in MAFLD patients.





Similar content being viewed by others
Abbreviations
- CAP:
-
Controlled attenuation parameter
- LSM:
-
Liver stiffness measurement
- ALT:
-
Alanine aminotransferase
- AST:
-
Aspartate aminotransferase
- BMI:
-
Body mass index
- BUN:
-
Blood urea nitrogen
- GGT:
-
γ-Glutamyl transferase
- ALP:
-
Alkaline phosphatase
- TB:
-
Total bilirubin
- LDL-C:
-
Low-density lipoprotein-cholesterol
- HDL-C:
-
High-density lipoprotein-cholesterol
- TG:
-
Triglyceride
- TC:
-
Total cholesterol
- SUA:
-
Serum uric acid
- PChE:
-
Pseudocholinesterase
- PLT:
-
Platelet count
- FBG:
-
Fasting blood glucose
- NAFL:
-
Nonalcoholic fatty liver
- NASH:
-
Nonalcoholic steatohepatitis
- MAFLD:
-
Metabolic associated fatty liver disease.
References
Ballestri S, Zona S, Targher G, Romagnoli D, Baldelli E, Nascimbeni F, Roverato A, Guaraldi G, Lonardo A. Nonalcoholic fatty liver disease is associated with an almost twofold increased risk of incident type 2 diabetes and metabolic syndrome. Evidence from a systematic review and meta-analysis. J Gastroenterol Hepatol. 2016;31(5):936–44.
Tarantino G, Citro V, Capone D. Nonalcoholic fatty liver disease: a challenge from mechanisms to therapy. J Clin Med. 2019;9(1):15.
Sanyal AJ. NASH: A global health problem. Hepatol Res. 2011;41(7):670–4.
Paredes-Turrubiarte G, Gonzalez-Chavez A, Perez-Tamayo R, Salazar-Vazquez BY, Hernandez VS, Garibay-Nieto N, Fragoso JM, Escobedo G. Severity of non-alcoholic fatty liver disease is associated with high systemic levels of tumor necrosis factor alpha and low serum interleukin 10 in morbidly obese patients. Clin Exp Med. 2016;16(2):193–202.
Gatselis NK, Ntaios G, Makaritsis K, Dalekos GN. Adiponectin: a key playmaker adipocytokine in non-alcoholic fatty liver disease. Clin Exp Med. 2014;14(2):121–31.
Eslam M, Newsome PN, Sarin SK, Anstee QM, Targher G, Romero-Gomez M, Zelber-Sagi S, Wai-Sun Wong V, Dufour JF, Schattenberg JM et al, A new definition for metabolic dysfunction-associated fatty liver disease: an international expert consensus statement. J Hepatol 2020.
Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64(1):73–84.
Rinella ME. Nonalcoholic fatty liver disease: a systematic review. JAMA. 2015;313(22):2263–73.
Nobili V, Alisi A, Valenti L, Miele L, Feldstein AE, Alkhouri N. NAFLD in children: new genes, new diagnostic modalities and new drugs. Nat Rev Gastroenterol Hepatol. 2019;16(9):517–30.
Arrese M, Cabrera D, Kalergis AM, Feldstein AE. Innate immunity and inflammation in NAFLD/NASH. Dig Dis Sci. 2016;61(5):1294–303.
Younossi ZM, Ratziu V, Loomba R, Rinella M, Anstee QM, Goodman Z, Bedossa P, Geier A, Beckebaum S, Newsome PN, et al. Obeticholic acid for the treatment of non-alcoholic steatohepatitis: interim analysis from a multicentre, randomised, placebo-controlled phase 3 trial. Lancet. 2019;394(10215):2184–96.
Neuschwander-Tetri BA, Loomba R, Sanyal AJ, Lavine JE, Van Natta ML, Abdelmalek MF, Chalasani N, Dasarathy S, Diehl AM, Hameed B, et al. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): a multicentre, randomised, placebo-controlled trial. Lancet. 2015;385(9972):956–65.
Hu YY, Dong NL, Qu Q, Zhao XF, Yang HJ. The correlation between controlled attenuation parameter and metabolic syndrome and its components in middle-aged and elderly nonalcoholic fatty liver disease patients. Medicine (Baltimore). 2018;97(43):e12931.
Brunt EM, Wong VW, Nobili V, Day CP, Sookoian S, Maher JJ, Bugianesi E, Sirlin CB, Neuschwander-Tetri BA, Rinella ME. Nonalcoholic fatty liver disease. Nat Rev Dis Primers. 2015;1:15080.
Karlas T, Petroff D, Sasso M, Fan JG, Mi YQ, de Ledinghen V, Kumar M, Lupsor-Platon M, Han KH, Cardoso AC, et al. Individual patient data meta-analysis of controlled attenuation parameter (CAP) technology for assessing steatosis. J Hepatol. 2017;66(5):1022–30.
Hyogo H, Yamagishi S, Maeda S, Fukami K, Ueda S, Okuda S, Nakahara T, Kimura Y, Ishitobi T, Chayama K. Serum asymmetric dimethylarginine levels are independently associated with procollagen III N-terminal peptide in nonalcoholic fatty liver disease patients. Clin Exp Med. 2014;14(1):45–51.
Eslam M, George J. Genetic contributions to NAFLD: leveraging shared genetics to uncover systems biology. Nat Rev Gastroenterol Hepatol. 2020;17(1):40–52.
Eslam M, George J. Genetic insights for drug development in NAFLD. Trends Pharmacol Sci. 2019;40(7):506–16.
Eslam M, Valenti L, Romeo S. Genetics and epigenetics of NAFLD and NASH: clinical impact. J Hepatol. 2018;68(2):268–79.
Moran M, Cheng X, Shihabudeen Haider Ali MS, Wase N, Nguyen N, Yang W, Zhang C, DiRusso C, Sun X. Transcriptome analysis-identified long noncoding RNA CRNDE in maintaining endothelial cell proliferation, migration, and tube formation. Sci Rep. 2019;9(1):19548.
Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12(12):861–74.
Ma S, Long T, Huang WJM: Noncoding RNAs in inflammation and colorectal cancer. RNA Biol 2019:1-8
Michalik KM, You X, Manavski Y, Doddaballapur A, Zornig M, Braun T, John D, Ponomareva Y, Chen W, Uchida S, et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res. 2014;114(9):1389–97.
Shi L, Hong X, Ba L, He X, Xiong Y, Ding Q, Yang S, Peng G. Long non-coding RNA ZNFX1-AS1 promotes the tumor progression and metastasis of colorectal cancer by acting as a competing endogenous RNA of miR-144 to regulate EZH2 expression. Cell Death Dis. 2019;10(3):150.
Li D, Cheng M, Niu Y, Chi X, Liu X, Fan J, Fan H, Chang Y, Yang W. Identification of a novel human long non-coding RNA that regulates hepatic lipid metabolism by inhibiting SREBP-1c. Int J Biol Sci. 2017;13(3):349–57.
Chen X, Xu Y, Zhao D, Chen T, Gu C, Yu G, Chen K, Zhong Y, He J, Liu S, et al. LncRNA-AK012226 is involved in fat accumulation in db/db mice fatty liver and non-alcoholic fatty liver disease cell model. Front Pharmacol. 2018;9:888.
Wang TH, Lin YS, Chen Y, Yeh CT, Huang YL, Hsieh TH, Shieh TM, Hsueh C, Chen TC. Long non-coding RNA AOC4P suppresses hepatocellular carcinoma metastasis by enhancing vimentin degradation and inhibiting epithelial-mesenchymal transition. Oncotarget. 2015;6(27):23342–57.
Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339(2):159–66.
Yan C, Chen J, Chen N. Long noncoding RNA MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability. Sci Rep. 2016;6:22640.
Chen G, Yu D, Nian X, Liu J, Koenig RJ, Xu B, Sheng L. LncRNA SRA promotes hepatic steatosis through repressing the expression of adipose triglyceride lipase (ATGL). Sci Rep. 2016;6:35531.
Li P, Ruan X, Yang L, Kiesewetter K, Zhao Y, Luo H, Chen Y, Gucek M, Zhu J, Cao H. A liver-enriched long non-coding RNA, lncLSTR, regulates systemic lipid metabolism in mice. Cell Metab. 2015;21(3):455–67.
Gong J, Qi X, Zhang Y, Yu Y, Lin X, Li H, Hu Y. Long noncoding RNA linc00462 promotes hepatocellular carcinoma progression. Biomed Pharmacother. 2017;93:40–7.
Cui H, Zhang Y, Zhang Q, Chen W, Zhao H, Liang J. A comprehensive genome-wide analysis of long noncoding RNA expression profile in hepatocellular carcinoma. Cancer Med. 2017;6(12):2932–41.
Wei Y, Hou H, Zhang L, Zhao N, Li C, Huo J, Liu Y, Zhang W, Li Z, Liu D, et al. JNKi- and DAC-programmed mesenchymal stem/stromal cells from hESCs facilitate hematopoiesis and alleviate hind limb ischemia. Stem Cell Res Ther. 2019;10(1):186.
Zhao Q, Zhang L, Wei Y, Yu H, Zou L, Huo J, Yang H, Song B, Wei T, Wu D, et al. Systematic comparison of hUC-MSCs at various passages reveals the variations of signatures and therapeutic effect on acute graft-versus-host disease. Stem Cell Res Ther. 2019;10(1):354.
Zhang X, Yang Y, Zhang L, Lu Y, Zhang Q, Fan D, Zhang Y, Zhang Y, Ye Z, Xiong D. Mesenchymal stromal cells as vehicles of tetravalent bispecific Tandab (CD3/CD19) for the treatment of B cell lymphoma combined with IDO pathway inhibitor D-1-methyl-tryptophan. J Hematol Oncol. 2017;10(1):56.
Zhang L, Wang H, Liu C, Wu Q, Su P, Wu D, Guo J, Zhou W, Xu Y, Shi L, et al. MSX2 initiates and accelerates mesenchymal stem/stromal cell specification of hPSCs by regulating TWIST1 and PRAME. Stem Cell Reports. 2018;11(2):497–513.
Zhang L, Liu C, Wang H, Wu D, Su P, Wang M, Guo J, Zhao S, Dong S, Zhou W, et al. Thrombopoietin knock-in augments platelet generation from human embryonic stem cells. Stem Cell Res Ther. 2018;9(1):194.
Zhang W, Liu C, Wu D, Liang C, Zhang L, Zhang Q, Liu Y, Xia M, Wang H, Su P, et al. Decitabine improves platelet recovery by down-regulating IL-8 level in MDS/AML patients with thrombocytopenia. Blood Cells Mol Dis. 2019;76:66–71.
Huo J, Zhang L, Ren X, Li C, Li X, Dong P, Zheng X, Huang J, Shao Y, Ge M, et al. Multifaceted characterization of the signatures and efficacy of mesenchymal stem/stromal cells in acquired aplastic anemia. Stem Cell Res Ther. 2020;11(1):59.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504.
Yao J, Chen N, Wang X, Zhang L, Huo J, Chi Y, Li Z, Han Z. Human supernumerary teeth-derived apical papillary stem cells possess preferable characteristics and efficacy on hepatic fibrosis in mice. Stem Cells Int. 2020;2020:6489396.
Brunt EM, Kleiner DE. Challenges in the hepatic histopathology in non-alcoholic fatty liver disease. Gut. 2017;66(9):1539–40.
Kleiner DE, Brunt EM, Wilson LA, Behling C, Guy C, Contos M, Cummings O, Yeh M, Gill R, Chalasani N, et al. Association of histologic disease activity with progression of nonalcoholic fatty liver disease. JAMA Netw Open. 2019;2(10):e1912565.
Middleton MS, Van Natta ML, Heba ER, Alazraki A, Trout AT, Masand P, Brunt EM, Kleiner DE, Doo E, Tonascia J, et al. Diagnostic accuracy of magnetic resonance imaging hepatic proton density fat fraction in pediatric nonalcoholic fatty liver disease. Hepatology. 2018;67(3):858–72.
Myers RP, Pollett A, Kirsch R, Pomier-Layrargues G, Beaton M, Levstik M, Duarte-Rojo A, Wong D, Crotty P, Elkashab M. Controlled Attenuation Parameter (CAP): a noninvasive method for the detection of hepatic steatosis based on transient elastography. Liver Int. 2012;32(6):902–10.
Liu K, Wong VW, Lau K, Liu SD, Tse YK, Yip TC, Kwok R, Chan AY, Chan HL, Wong GL. Prognostic value of controlled attenuation parameter by transient elastography. Am J Gastroenterol. 2017;112(12):1812–23.
Petta S, Di Marco V, Pipitone RM, Grimaudo S, Buscemi C, Craxi A, Buscemi S. Prevalence and severity of nonalcoholic fatty liver disease by transient elastography: genetic and metabolic risk factors in a general population. Liver Int. 2018;38(11):2060–8.
Zhao XY, Xiong X, Liu T, Mi L, Peng X, Rui C, Guo L, Li S, Li X, Lin JD. Long noncoding RNA licensing of obesity-linked hepatic lipogenesis and NAFLD pathogenesis. Nat Commun. 2018;9(1):2986.
Di Mauro S, Scamporrino A, Petta S, Urbano F, Filippello A, Ragusa M, Di Martino MT, Scionti F, Grimaudo S, Pipitone RM, et al. Serum coding and non-coding RNAs as biomarkers of NAFLD and fibrosis severity. Liver Int. 2019;39(9):1742–54.
Acknowledgements
The work was supported by the National Natural Science Foundation of China (81660410), Natural Science Foundation of Tianjin (19JCQNJC12500), Science and Technology Project of Tianjin (17ZXSCSY00030), Project funded by China Postdoctoral Science Foundation (2019M661033), Natural Science Foundation of Yunnan (2017FE468-174) and Yunnan Science and Technology Project (2015HB073, CXTD201610, L-2017018, 2018NS0100), Nanyang Science and Technology Project of He-nan Province (JCQY012), Key project funded by Department of Science and Technology of Shangrao City (2020, to ZCH), the Reserve Training Project of "Thousand" Project of Health Science and Technology Talents in Kunming (2019-sw-52), NHC Key Laboratory of Drug Addiction Medicine (Kunming Medical University). The authors thank all the nurses and patients involved in this study. We also thank the precision medicine division of Health-Biotech (Tianjin) Stem Cell Research Institute Co., Ltd. and the enterprise postdoctoral working station of Tianjin Chase Sun Pharmaceutical Co., Ltd. for their technical support.
Funding
The work was supported by the National Natural Science Foundation of China (81660410), Natural Science Foundation of Tianjin (19JCQNJC12500), Science and Technology Project of Tianjin (17ZXSCSY00030), Project funded by China Postdoctoral Science Foundation (2019M661033), Natural Science Foundation of Yunnan (2017FE468-174) and Yunnan Science and Technology Project (2015HB073, CXTD201610, L-2017018, 2018NS0100), Nanyang Science and Technology Project of He-nan Province (JCQY012), Key project funded by Department of Science and Technology of Shangrao City (2020, to ZCH), the Reserve Training Project of "Thousand" Project of Health Science and Technology Talents in Kunming (2019-sw-52), NHC Key Laboratory of Drug Addiction Medicine (Kunming Medical University).
Author information
Authors and Affiliations
Contributions
HY and QL contributed to collection and assembly of data, manuscript writing; LZ, MZ, JN, FX, LY, QQ, YL, ZD and CX contributed to collection and assembly of data; and HY, LZ and KW contributed to conception and design, data analysis and interpretation, manuscript writing, final approval of manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests and all authors consent to publish the data.
Availability of data and material
All data generated or analyzed during this study are included in this published article and its supplementary information files. Meanwhile, the datasets used and analyzed during the current study are also available from the corresponding author on reasonable request.
Code availability
Not applicable.
Ethics approval and consent to participate
The study was performed in accordance with the principles set by the Declaration of Helsinki, and was approved by the ethics committee of First Affiliated Hospital of Kunming Medical University (approval number: 2020-L-08). Informed consent was obtained from all individual participants included in the study.
Consent for participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yang, H., Li, Q., Zhang, L. et al. LncPRYP4-3 serves as a novel diagnostic biomarker for dissecting subtypes of metabolic associated fatty liver disease by targeting RPS4Y2. Clin Exp Med 20, 587–600 (2020). https://doi.org/10.1007/s10238-020-00636-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10238-020-00636-1