Abstract
This paper provides first information on organ-like bacterial aggregates in the tentacles of the sea anemone Metridium senile. The specimens were collected from waters near Helgoland (German Bight, North Sea) and the Orkney Islands. Tentacles were prepared for morphological inspection by light and scanning electron microscopy as well as for the phylogenetic analysis of endocytic bacteria. Bacterial aggregates are located in caverns of the tentacles’ epidermis. The aggregates are enwrapped in thin envelopes, which contain coccoid and/or rod-shaped tightly packed bacteria of different division states. Most of the bacterial cells are connected by fine filamentous structures. The phylogenetic determination is based on the sequence data of the 16S rDNA derived from tentacle material. Sequence analysis revealed three different subgroups of intratentacular proteobacteria. The dominant band, detected in all of the samples tested, showed a close relationship (98%) to a gram-negative Endozoicimonas elysicola. Two bands, only detected in tentacles of M. senile from Helgoland were assigned to Pseudomonas saccherophilia (99%), a knallgas bacterium, and to Ralstonia pickettii (100%). The bacteria represent a specific bacterial community. Their DGGE profiles do not correspond to the profiles of the planktonic bacteria generated from seawater close to the habitats of the anemones. The allocation of DNA sequences to the different morphotypes, their isolation, culturing and the elucidation of the physiological functions of intratentacular bacteria are in progress.
Similar content being viewed by others
Introduction
The fascinating interactions between bacteria and invertebrates are common in marine habitats. However, most of their ecological functions are still enigmatic. Current literature describes endobiotic bacteria in various eukaryotic host organisms such as protozoans, sponges, cnidaria, annelids, echinoderms, and ascidians (Deming and Colwell 1982; Paul et al. 1986; Cary et al. 1997; Burnett and McKenzie 1997; Althoff et al. 1998; Groepler and Schuett 2003). However, the key questions concerning the phylogenetic relationship of bacterial symbionts (Kirchner et al. 1999; Seibold et al. 2001; Moss et al. 2003; Wichels et al. 2004; Schuett et al. 2005), their ecological and physiological function, their secondary metabolites, their chemical structure and the effects of these compounds on host tissue, are mostly cryptic. Apart from floating or motile endocytic bacteria in the cytosol often detected in the tissue material, a higher form of bacterial organization exists. This comprises specific cyst-like bacterial aggregates, also known as bacteriocytes. They are common in various orders of insects (e.g. Homoptera, Hymenoptera, Blattaria, Coleoptera). Those endosymbiotic bacteria belong to genera such as Buchnera, Wolbachia, Wigglesworthia, Candidatus Blochmannia (van Ham et al. 2003; Gomez-Valero et al. 2004; Wolschin et al. 2004). They may have vital functions for host growth or may play additional nutritional roles (Wernegreen et al. 2002; Gill et al. 2003). Bacteriocytes are also common in the gut region and the ovaries of the ant Camponotus floridanus.The very few reports on bacteriocyte-like structures in marine invertebrates describe bacterial aggregates located in the gills of the bivalve Loripes lucinalis. These endocellular symbiotic bacteria have the ability to oxidize sulphur (Southward 1986; Johnson and Fernandez 2001). Concerning the phylum cnidaria, Palincsar et al. (1988, 1989) provided the only reports on bacteriocyte-like structures in the epidermis of the sea anemone Aiptasia pallida. Fatty acid profiles suggested that the bacteria might belong to the genus Vibrio. This anemone inhabits subtropical regions of the Atlantic Ocean and does not inhabit colder waters. A. pallida harbors algal symbionts in the gastrodermis. The present report provides first morphological and phylogenetic information on organ-like intratentacular bacterial aggregates in the sea anemone Metridium senile.
Materials and methods
Samples and preparation
Individuals of M. senile were collected from the shallow waters around Helgoland and from waters around the Orkney Islands by divers of the Biologische Anstalt Helgoland. During the investigation the anemones were kept at ambient temperature in aquaria which contained seawater enriched with natural plankton. To prevent the animals from contracting they were anaesthetized with MgCl2 (3% final conc.). Tentacles were clipped off and subjected to washing procedures in order to remove potentially contaminating epibiotic bacteria. Samples were washed twice in sterile seawater for 5 min, followed by an additional 5 min treatment in cetyl-trimethyl-ammonium-bromide (CTAB; final concentration 10 μg/ml seawater). In a subsequent step CTAB was washed away three times with sterile seawater (5 min each). After removing the seawater, M. senile samples were directly used for further procedures. Light microscopic and scanning electron microscopic controls showed no bacteria attached to the surface of fresh unwashed tentacle material of M. senile.
Microscopic preparation
Light microscopy For inspection tentacles were transferred to microscopic slides. After reducing salinity (presence of distilled water enhanced the release of the bacterial aggregates), tentacles were carefully squeezed between coverslip and slide until the bacterial aggregates became sufficiently translucent for microscopic investigation.
Scanning electron microscopy (SEM) The preparation comprised the following steps: (1) fixation of tentacle material in 4.0% glutaraldehyde, Na–K–phosphate buffer PBS (0.1 M) at pH 7.0 for 2 h. (2) Replacing seawater by ethanol in steps of 30, 50, 70, 80, 90 and 96%. (3) Ethanol exchange by amyl acetate in steps of 25, 50, 75 and 100%. (4) Critical point drying (Bal-Tec) in liquid CO2 at a pressure of 72.9 bar and 31.1°C. (5) Mounting of tentacle samples. (6) Au-coating at 60 mA for 90 s (sputtering system SCD 030, Balzers). In order to allow insight into the inner location of the bacterial aggregates, tentacle tips were snapped off and the resulting cutting areas were gold coated. (7) Samples were investigated with a SEM field emission scanning microscope (S-800, Hitachi).
Direct PCR amplification of 16S rDNA fragments
As intratentacular bacterial DNA concentration is too low for DNA extraction, prokaryotic DNA was directly amplified without prior extraction. Bacterial aggregates were released from tentacles in the presence of distilled water and successively sampled by the use of sterilized long human eyelashes functioning as effective collecting tools. Bacterial aggregates were then ready for further processing. The complete procedures of sample treatment including deproteination steps, freeze–thaw-lysing-treatment, Lyse-N-Go-treatment (Pierce) and PCR parameters (primers 341fc, 5′-cgc ccg ccg cgc ccc gcg ccc ggc ccg ccg ccc ccg ccc ccc tac ggg agg cag cag-3′, clamp region underlined; 907rwob, 5′-ccg tca att cct ttr agt tt-3′; Muyzer et al. 1995) have been described by Schuett et al. (2005). Negative controls were carried out without template DNA; Escherichia coli J53 served as positive control.
DGGE analysis of PCR products
Prior to DGGE the quantity of amplified PCR products was determined by analyzing 5 μl PCR product on 1.2% agarose gels. Bands stained with ethidium bromide were visualized by using a transilluminator (Pharmacia) and documented with a MP-4 camera (Polaroid Corp.).
DGGE with prokaryotic PCR samples (30–60 μl) was performed by using the DCode electrophoresis system (Biorad). Preparation of polyacrylamide gels (15–70% denaturant gradient) and electrophoresis parameters (at 100 V for 15 h) were performed according to Muyzer et al. (1995). Bands were stained with SYBR-gold (documentation see above) and excised from polyacrylamide gels. DNA was extracted from gel material and dissolved in 10 μl d. w. (Sambrook et al. 1989).
Re-amplification of DNA fragments from DGGE bands
A further purification of DNA fragments could be obtained by re-amplifying the extracted DNA using again the forward primer with clamp (341fc) and checking the exact position of the band after an additional DGGE run (30–45; 35–55% denaturant gradient). These bands were excised again and re-amplified for sequencing, this time applying the forward primer without clamp (341f). The components of PCR cocktails (100 μl) have been described completely by Schuett et al. (2005). PCR products were purified by using the Qiaquick PCR Purification Kit (Qiagen) following the instructions of the manufacturer’s protocol and eluted with 50 μl d. w. Quantities of purified DNA samples were determined by electrophoresis (1.2% agarose gels).
DNA sequencing of PCR products and comparative sequence analysis
Sequencing was carried out by Qiagen Sequencing Services/Hilden, Germany.
Sequences were aligned by using the Align IR program and the advanced BLAST search program of the National Center of Biotechnology Information (NCBI) website (http://www.ncbi.nlm.nih.gov/Blast) to find closely related sequences. Data were screened for existing chimera by applying the “Ribosomal Database Project” (http://www.rdp.cme.msu.edu/cgis/chimera.cgi).
Bacterioplankton of the surrounding seawater
For a comparison of intratentacular bacteria with the planktonic bacteria, surface water samples (1 m depth) from Helgoland Roads were collected, and two filter fractions (3–60 and 0.2–3 μm) from the surface water samples were prepared. DNA extractions were performed according to Anderson and Mc Kay (1983) protocol omitting as well the DNA denaturing step with sodium hydroxide as the latter neutralization step. Further processing of the extracts including PCR amplification, DGGE and sequencing has been accomplished according to the measures described above.
Results
Microscopic investigation
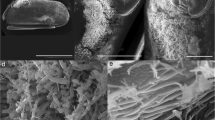
Bacterial aggregates were carefully collected with eyelashes. The aggregates containing the bacteria were documented by light microscopy (Figs. 1, 2). The scanning electron microscopic investigation revealed that the bacterial aggregates resemble organs, which were located in caverns of the tentacle’s epidermis (Figs. 3, 4). Aggregates of 30-μm diameter may contain an estimated number of 13,000 bacterial cells. The number of aggregates varies between a few and more than ten per tentacle. High numbers of aggregates were usually found in the tip region of the tentacles. The active bacteria with different division states were connected by filamentous structures and wrapped in thin and fragile envelopes (Fig. 5). The gram-negative rods (according to the gram test) were tightly packed as bacteria displaying fingerprint-like structures of diameters between 10 and 100 μm. Their length was between 1 and 2 μm with a width of 0.5 μm of rod-shaped bacteria. No morphological differences were found between bacterial aggregates from the tentacles of the M. senile collected near Helgoland and those from the Orkney Islands. The tentacles disintegrated after some hours; thereby the aggregates were extruded from tissue material. The thin envelopes opened up and the non-motile individual bacterial cells were released from the tightly packed aggregates.
DGGE profiles of intratentacular bacteria and their phylogenetic affiliation
Sample material comprised bacterial aggregates from tentacles of M. senile. Bacterial 16S rDNA from tentacles was directly amplified by PCR. DGGE profiles showed ca. four distinguishable OTUs. Figure 6 shows the DGGE profiles of six samples; three samples from each of the locations at Helgoland and the Orkney Islands. The bands were excised, re-amplified and successfully sequenced. The sequence data suggest the presence of three different proteo-bacterial sub-groups in the sample material. The relationship of the bacteria identified is close (89–100%) to those described in literature. The dominant bands (# 265 and # 280) were detected in all of the samples from both Helgoland and the Orkney Islands. The fragments tested displayed a close relationship of 98% to Endozoicimonas elysicola (γ-subgroup of proteobacteria), representing a novel species of a novel genus (Kurahashi and Yokoto 2006). Weaker bands, located well above the main bands discussed earlier, were found in all of the six samples. As a representative of this group of weak bands, fragment # 276 was affiliated also with a relationship of 98% to E. elysicola. Fragments only found in M. senile from Helgoland were the following: a solitary band # 275 was affiliated to Pseudomonas saccherophilia (99%), a knallgas bacterium and the band # 269 could be allocated to Ralstonia pickettii (100%). The DGGE profiles of bacterioplankton from Helgoland Roads exhibited no similarity to the DGGE profiles of intratentacular bacteria of M. senile from the same location (Fig. 7).
DGGE-profiles (15–70% denaturant gradient) of bacterial communities in tentacles of M. senile from coastal waters of Helgoland and Orkney Islands assigned to species described in literature. Marked bands (selected DNA-fragments were re-amplified and sequenced) represent next neighbors to Endozoicimonas elysicola (bands # 265, # 276, # 280); Pseudomonas saccharophilia (# 275); Ralstonia pickettii (# 269)
Comparison between DGGE profiles (30–45% denaturant gradient) of planktonic bacteria from Helgoland Roads (1 m depth) and intratentacular bacteria from M. senile. A Planktonic bacteria (filter fraction <3.0–>0.2 μm), lanes 1–3. B Intratentacular bacteria from M. senile, collected from waters of Helgoland and the Orkney Islands: lanes 4–8; lane 4 harbors band # 265; lane 7 band # 276 and lane 8 band # 280, all three bands represent Endozoicimonas elysicola; lane 5 harbors band # 269 representing Ralstonia pickettii; lane 6 harbors band # 275 (located out of gel range) representing Pseudomonas saccharophilia detected in a 35–55% denaturant gradient. C Planktonic bacteria (filter fraction <60–>3.0 μm), lanes 9–11
Discussion
Palincsar et al. (1989) provided the only report on cyst-like bacterial aggregates in the epidermis of the sea anemone A. pallida. The comparison between the bacterial aggregates detected in the A. pallida species of the warmer Atlantic regions which additionally harbour algal symbionts and M. senile, a typical species of the boreal environment of the North Sea and the northern Atlantic, unveiled a series of similarities and some noticeable differences: the main similarities comprise the various numbers of aggregates in the tentacles, the morphological fingerprint structures at microscopic inspection when sample material was squeezed between slide and cover slip, the size of aggregates (diameter between 10 and 100 μm), their location in the tentacle’s epidermis, and finally, the thin envelope covering the bacterial aggregates. Noticeable differences mainly refer to the identification of the endobiotic bacteria. Palincsar et al. (1989) identified the intratentacular bacteria of A. pallida as members of the genus Vibrio; these data were based on fatty acid analysis. The present phylogenetic analysis strongly suggests that the bacteria embedded in aggregates are closely related to Endozoicimonas elysicola. This organism was found to be a symbiont of the tropical nudibrach Elysia ornata. Bowman and Nowak (2004) suspect its role in amoebic fish disease. Other differences refer to findings that the bacteria of A. pallida are not connected by filamentous structures, and that in contrast to A. pallida the stems of M. senile do not harbour aggregates.
Pseudomonas saccherophilia and Ralstonia pickettii, however, were detected only in the tentacles derived from the Helgoland sample material, a major difference between the sample material from Helgoland and the Orkney Islands. Both of these organisms seem to be more common as endobiotic bacteria. They were previously detected in the ascidian tunic matrix of Diplosoma migrans collected from waters of Helgoland (Schuett et al. 2005). Pseudomonas saccherophilia is known as “knallgas bacterium” with a specific ability to hydrolyse starch and gelatin (Aragno and Schlegel 1992). Ralstonia pickettii (Coenye et al. 2003) is known as a pathogenic organism of clinical sources. At the present state these data do not allow a reliable interpretation of the ecological functions. Nevertheless, the data strongly suggest that intratentacular bacteria represent a specific bacterial community, which is underlined by the fact that the DGGE profiles of intratentacular bacteria do not correspond to the profiles of the planktonic bacteria from the surrounding seawater. In order to reach a broader separation distance, changes of the urea gradient have been helpful. The use of nearest neighbour data analysis in combination with prokaryotic 16S rDNA fragments of bacteria embedded in eukaryotic tissue material is often problematic: direct PCR is not always successful. Furthermore bands of the same sequence may occur at different positions during first separation. Figure 6 shows that band # 276 is located in the upper gel part of the denaturant gradient 15–70%. A second gel run (altered gradient 30–45%) with the re-amplified DNA material shifts the band # 276 to a lower location, matching the position of band # 280 (Fig. 7). Sequence data here demonstrate that both bands represent the same organism. This observation allows the conclusion that both fragments are composed of the same base sequences; however, both these fragments must possess different steric structures, which are responsible for the variable migration behavior. Obstacles occur if DGGE bands are too closely located. Some major questions are still open: the allocation of bacterial morphotypes to bands is unclear. Origin and transfer mechanisms of the aggregate-bacteria are not definite. Ecological function of these bacteria is unknown. The mode of interaction—symbiosis versus parasitism—is currently not understood. Postulating a symbiotic process, can the organ-like aggregates be regarded as a stage in evolution? Likewise, the function(s) of filamentous structures connecting aggregate bacteria is enigmatic. Do these connecting structures serve as communicating medium between the bacteria or do they simply have mechanical stabilizing function? Finally, the chemical structure of the envelopes covering the aggregates is open to question. Do they function as membranes, and are they produced by the eukaryotic host or the bacterial tenants?
References
Althoff K, Schuett C, Krasko A, Steffen R, Batel R, Müller WEG (1998) Evidence for symbiosis between bacteria of the genus Rhodobacter and the marine sponge Halichondria panicea: harbour also putatively-toxic bacteria? Mar Biol 130:529–536
Anderson DG, Mc Kay LL (1983) Simple and rapid method for isolation of large plasmidDNA from lactic streptococci. Appl Environ Microbiol 46:549–552
Aragno M, Schlegel HG (1992) The mesophilic hydrogen-oxidizing (knallgas) bacteria. In: Ballows A, Trüper HG, Dworkin M, Harder W, Schleifer KH (eds) The prokaryotes, vol 1, 2nd edn. Springer, New York, pp 344–384
Bowman JP, Nowak B (2004) Salmonid gill bacteria and their relationship to amoebic gill disease. J Fish Dis 27:483–492
Burnett WJ, McKenzie JD (1997) Subcuticular bacteria from the brittle star Ophiactis balli (Echinodermata, Ophiuroidea) represent a new lineage of extracellular marine symbionts in the α-subdivision of the class proteobacteria. Appl Environ Microbiol 63:1721–1724
Cary SC, Cottrell MT, Stein JL, Camacho F, Desbueres D (1997) Molecular identification and location of filamentous symbiotic bacteria associated with hydrothermal vent annelid Alvinella pompejana. Appl Environ Microbiol 63:1124–1130
Coenye T, Goris J, De Vos P, Vandamme P, LiPuma J (2003) Classification of Ralstonia pickettii isolates from the environment and clinical samples as Ralstonia insidiosa. Int J Syst Evol Microbiol 53:1075–1080
Deming J, Colwell RR (1982) Barophilic bacteria associated with digestive tracts of abyssal holothurians. Appl Environ Microbiol 44:1222–1230
Gill R, Silva FJ, Zientz E, Delmotte F, González-Candelas F, Latorre A, Rausell C, Kamerbeek J, Gadau J, Hölldobler B, van Ham RCHJ, Gross R (2003) The genome sequences of Blochmannia floridanus: comparative analysis of reduced genomes. PNAS 100(16):9388–9393
Gomez-Valero L, Soriano-Navarro M, Pérez-Brocal V, Heddi A, Moya A, Garciá-Verdugo JM, Latorre A (2004) Coexistence of Wolbachia with Buchnera aphidicola and a secondary symbiont in the aphid Cinara cedri. J Bacteriol 186:6626–6633
Groepler W, Schuett C (2003) Bacterial community in the tunic matrix of a colonial ascidian Diplosoma migrans. Helgol Mar Res 57:139–143
van Ham RCHJ, Kamerbeek J, Palacios C, Rausell C, Abascal F, Bastolla U, Fernández JM, Jiménez L, Postino M, Silva JF, Tamanes J, Viguera E, Latorre A, Valencia A, Morán F, Moya A (2003) Reductive genome evolution in Buchnera aphidicola. PNAS 100(2):581–586
Johnson MA, Fernandez C (2001) Bacterial symbiosis in Loripes lucinalis (Mollusca: Bivalvia) with comments on reproductive strategy. J Mar Biol Ass UK 81:251–257
Kirchner M, Sahling G, Schuett C, Doepke H, Uhlig G (1999) Intracellular bacteria in the red tide-forming heterotrophic dinoflagellate Noctiluca scintillans. Arch Hydrobiol Spec Issues Adv Limnol 54:297–310
Kurahashi M, Yokoto A (2006) Endozoicimonas elysicola gen. nov., sp. nov., a γ-proteobacterium isolated from the sea slug Elysia ornata. Syst Appl Microbiol (Epub doi:10.1016/j.syapm.200607.003)
Moss C, Green DH, Perez B, Velasco A, Henriquez R, McKenzie JD (2003) Intracellular bacteria associated with the ascidian Ecteinascidia turbinata: phylogenetic and in situ hybridization analysis. Mar Biol 143:99–110
Müller WA, Leitz T (2002) Metamorphosis in the cnidaria. Can J Zool 80:1755–1771
Muyzer G, Hottenträger S, Teske A, Wawer C (1995) Denaturing gradient gel electrophoresis of PCR-amplified 16S rDNA. A new molecular approach to analyze the genetic diversity of mixed microbial communities. Mol Microb Ecol Manual 3.44, pp 1–22
Palincsar EE, Jones WR, Palincsar JS, Glogowski MA, Mastro JL (1989) Bacterial aggregates within the epidermis of the sea anemone Aiptasia pallida. Biol Bull 177:130–140
Paul JH, De Flawn MF, Jeffrey WH (1986) Elevated levels of microbial activity in the corral surface microlayers. Mar Ecol Progr Ser 33:29–40
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning. A laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Schuett C, Doepke H, Groepler W, Wichels A (2005) Diversity of intratunical bacteria in the tunic matrix of the colonial ascidian Diplosoma migrans. Helgol Mar Res 59:136–140
Seibold A, Wichels A, Schuett C (2001) Diversity of endocytic bacteria in the dinoflagellate Noctiluca scintillans. Aquat Microb Ecol 25:229–235
Southward (1986) Gill symbionts in Thyasirids and other bivalve molluscs. J Mar Biol Ass UK 81(66):889–914
Wernegreen JJ, Lazarus AB, Degnan PH (2002) Small genome of Candidatus blochmannia, the bacterial endosymbiont of Camponotus, impies irreversible specialization to an intracellular lifestyle. Microbiology 148:2551–2556
Wichels A, Hummert C, Elbrächter M, Luckas B, Schuett C, Gerdts G (2004) Bacterial diversity in toxic Alexandrium tamarense blooms off the Orkney Isles and the Firth of Forth. Helgol Mar Res 58:93–103
Wolschin F, Hölldobler B, Gross R, Zientz E (2004) Replication of endosymbiotic bacterium Blochmannia floridanus is correlated with the developmental and reproductive stages of its ant host. Appl Environ Microbiol 70:4096–4102
Acknowledgments
We are grateful to the BAH divers group who provided excellent fresh cnidarian sample material. Special thanks go to Dr. Karl Herrmann (University of Erlangen-Nürnberg) for helpful hints regarding the handling of animals and the preparation of the bacterial aggregates.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H.-D. Franke.
Rights and permissions
About this article
Cite this article
Schuett, C., Doepke, H., Grathoff, A. et al. Bacterial aggregates in the tentacles of the sea anemone Metridium senile . Helgol Mar Res 61, 211–216 (2007). https://doi.org/10.1007/s10152-007-0069-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10152-007-0069-4