Abstract
The purpose of this systematic review was to compare the diagnostic ability of blood markers for colorectal cancer (CRC). A systematic review of the literature for diagnostic blood markers for primary human colorectal cancer over the last 5 years was performed. The primary outcome was to assess the diagnostic ability of these markers in diagnosing colorectal cancer. The secondary outcome was to see whether the marker was compared to other markers. The tertiary outcome was to assess diagnostic ability in early versus late CRC, including stage IV disease. We identified 51 studies (29 prospective, 14 retrospective, and 8 meta-analyses). The markers were divided in broadly four groups: nucleic acids (RNA/DNA/messenger RNA/microRNAs), cytokines, antibodies, and proteins. The most promising circulating markers identified among the nucleid acids were NEAT_v2 non-coding RNA, SDC2 methylated DNA, and SEPT9 methylated DNA. The most promising cytokine to detect CRC was interleukin 8, and the most promising circulating proteins were CA11-19 glycoprotein and DC-SIGN/DC-SIGNR. Sensitivities of these markers for detecting primary colorectal carcinoma ranged from 70 to 98% and specificities from 84 to 98.7%. The best studied blood marker was SEPT9 methylated DNA, which showed great variability with sensitivities ranging from 48.2 to 95.6% and specificities from 80 to 98.9%, making its clinical applicability challenging. If combined with fecal immunochemical test (FIT), the sensitivity improved from 78 to 94% in detecting CRC. Methylated SEPT9, methylated SDC2, and -SIGN/DC-SIGNR protein had better sensitivity and specificity than CEA or CA 19-9. With the exception of SEPT9 which is currently being implemented as a screening test for CRC all other markers lacked reproducibility and standardization and were studied in relatively small population samples.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
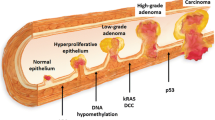
Colorectal cancer (CRC) is the third commonest cancer worldwide and caused 15,903 deaths in 2014 in the United Kingdom alone [1]. The stage of disease at diagnosis is the most important factor dictating survival. If the cancer is detected early, the reported 5-year survival rate is 90%, which can decrease to 14% if the disease is advanced on diagnosis [2]. The natural history of CRC is to develop from a benign adenoma, and the estimated time interval for development from normal mucosa to adenoma to invasive adenocarcinoma is 5–10 years [3, 4]. Detecting the disease early, therefore, is key to reducing mortality.
As most patients with CRC are asymptomatic or have non-specific symptoms in the early stages, it is vital to find a safe, acceptable, sensitive, specific, and cost-effective test that detects the early stage of the disease [5].
Currently, colonoscopy is the gold-standard diagnostic test to identify colonic pathology [2, 6]. A meta-analysis in 2015 by Brenner et al., showed that colonoscopy is estimated to reduce colorectal cancer incidence by 69% and mortality by 68% [7]. However, this is invasive, has low adherence, and is associated with potential risks to the patient [2]. Its alternative, virtual colonoscopy still requires bowel preparation and can cause discomfort to the patient. The risk of unnecessary radiation especially in the young is also an important disadvantage [8]. Other screening investigations include flexible sigmoidoscopy, fecal occult blood test (FOBT), and fecal immunohistochemistry test (FIT), which is a DNA-based fecal test. Although the FOBT has less than 50% sensitivity for CRC, the FIT has a reported 78% sensitivity and 96% specificity [9]. Aversion to handling stool is an important reason for the low uptake of the test. Only 58% of patients who are sent the FOBT return a sample. In the UK, the introduction of the DNA-based fecal test, which is easier to use, is expected to increase uptake to 75% by 2020, with the challenge of obtaining a sample for testing from stool still remaining [10].
Blood-based markers in current use, such as carcinoembryonic antigen (CEA) and cancer antigen (CA) 19-9, are for surveillance and for monitoring response to treatment but have a low sensitivity and specificity ranging from 40 to 70% and 73 to 90%, respectively, making them unsuitable as screening or diagnostic markers [5, 10] A more recently proposed marker, which is commercially available, is methylated septin 9. This is a molecular-based blood test whose reported accuracy in the literature has been variable. A recent meta-analysis by Yan et al. has shown sensitivity of 76% and specificity 87% making it comparable to the DNA-based fecal test [11].
The purpose of this study is a systematic review of the literature on diagnostic biomarkers in blood or tissue over the last 5 years in colorectal cancer.
Materials and methods
Inclusion criteria
-
Primary human studies and meta-analyses in the last 5 years, which assessed serum or tissue markers’ diagnostic ability in CRC.
Exclusion criteria
-
Studies looking at familial or inherited CRC
-
Studies with less than 100 patients overall
-
Animal or in vitro studies
-
Studies that did not specify sensitivity or specificity of the markers.
Search strategy
An electronic search of PubMed, EMBASE, Cochrane, and ISI Web of Science was performed for the relevant studies between January 2013 and December of 2017 using the following terms: (marker OR biomarker) AND (serum OR blood OR tissue) AND (diagnosis OR screening) AND (colorectal OR colon OR bowel or rectal) AND (cancer OR carcinoma OR neoplasia). There were no language restrictions and duplicates were removed. After reviewing the title and abstract of the studies, as per the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) flowchart, the relevant manuscripts were selected for full-text review (Fig. 1). One additional relevant article was added. A 5-year timeframe was chosen to provide a summary of the recent advances in biomarkers and of the ones that have potential for future use.
Data extraction
All original studies and meta-analyses which assessed diagnostic ability of markers were included. Data extracted from each full-text manuscript were as follows: authors, publication year, whether it was serum or plasma or whole blood or tissue, area under the curve, sensitivity, and specificity of detecting primary CRC and if compared to another standard test and cutoff values. If the AUC or sensitivity or specificity was not specifically determined the study was excluded from the analysis.
Study outcomes
The primary outcome was to compare the diagnostic ability of the markers studied in blood or tissue. The secondary outcome was to see whether the marker was compared to other markers. The tertiary outcome was to assess the diagnostic ability of the marker in early versus late CRC.
Results
The literature search over the last 5 years yielded 2844 papers from which 51 studies were eligible for inclusion in the review (Fig. 1).
The markers were divided in broadly four groups: nucleic acids (RNA/DNA/messenger RNA/microRNAs), cytokines, antibodies, and proteins. The nucleic acid category was further subdivided into single microRNA, panel of microRNAs, and a separate group with RNA, DNA, and messenger RNA. They are summarized in Tables 1, 2, 3, 4, 5 and 6. Overall, 29 prospective studies, 14 retrospective studies, and 8 meta-analyses were included.
Nucleic acids (Tables 1, 2, 3)
MicroRNAs (Tables 1, 2)
These are small non-coding RNA particles, which regulate gene expression by binding to messenger RNA (mRNA) and affecting protein translation or gene expression. They are thought to act as either tumor suppressor genes or oncogenes. In the literature, they have been investigated singly or in panels to assess their diagnostic and prognostic capabilities. A study done by Imaoka et al. in 2016, reported that mi-1290 showed promise as a diagnostic marker. Mi-1290, is thought to promote epithelial mesenchymal transition (EMT), proliferation and has metastatic potential. The study investigated 324 patients and showed a sensitivity of 70.1% and specificity of 91.2% in detecting CRC. Its sensitivity in detecting adenomas compared to controls was 46.4% and specificity 91.2%, therefore inadequate to be used as a screening test on its own [11].
Table 1 shows studies that investigated a single miRNA in colorectal cancer and Table 2 shows the studies that investigated panels of these markers.
A meta-analysis performed by Zhang et al. for mi-21 showed a sensitivity and specificity of 81% [12] whilst Xu F et al.’s meta-analysis for the same marker in 2015, showed a pooled sensitivity of 75% with specificity of 84%. Among the original studies, the most promising was for mi-1290, which showed an AUC of 0.830, sensitivity of 70.1% and specificity of 91.2% [11].
Assessing different combinations of miRNAs had variable results. Fang et al. investigated a panel of three markers (mi-24, mi-320a, and mi-423-5p) which showed an overall sensitivity of 92.79% and specificity of 70.77% in detecting CRC and also showed high sensitivity and specificity in detecting early cancer [13].
RNA/DNA/messenger RNA (Table 3)
Septin 9, which has been well studied and is now commercially available [Epi proColon 2.0 (Epigenomics), mS9 (Abbott Molecular), ColoVantage (Quest Diagnostics)], has had disputed results. A meta-analysis by Nian et al. published in 2017, included 25 studies, of which only 2 showed a low risk of bias. Twenty-one studies excluded “difficult-to-diagnose” patients and seven studies did not specify thresholds used. The pooled sensitivity was 72% and specificity of 92% which if combined with FIT can increase up to 94% sensitivity, with a decreased specificity of 68% [14]. It also highlighted that the sensitivity for stage I disease was 45% and for polyps was 15% which makes it rather poor for a screening test.
A prospective analysis by Wu et al. investigated long non-coding RNA nuclear-enriched abundant transcript variants 1 and 2 (NEAT_v1 and NEAT_v2). Non-protein coding RNAs are greater than 200 nucleotides and constitute more than 70% of the genome. Non-protein coding RNA nuclear-enriched abundant gene 1 has 2 transcripts: NEAT1_v1 and NEAT1_v2. NEAT1_v2 showed a 70% overall sensitivity and 96% specificity in detecting CRC from controls, although the mean value in early versus late CRC was not significantly different [15]. Further study with larger cohorts and in different types of cancer is required to further validate this marker.
Hao et al., investigated ALU sequences in circulating free DNA, which are the most active sequences in the human genome [16]. ALU115, ALU247/115, and CEA, had a sensitivity of 85.57% and specificity of 97.27% in detecting CRC [16].
SDC2 is an integral membrane protein and is known to participate in cell migration and proliferation of cells. The SDC2 gene is expressed in mesenchymal but not epithelial colonic cells. It is also expressed in pancreatic epithelial cells. SDC2 methylation of DNA in a prospective study by Oh T et al. in 2013 showed sensitivity of detecting early CRC of 92% although this needs further validation in a larger study [17].
Messenger RNA conveys information from DNA to protein products of the genes expressed. The studies looking into mRNA have many flaws in the study design and description of control groups and many do not investigate the sensitivity and specificity of the marker in detecting early CRC. Cyclin E, p27kipl, and ki-67, investigated by Li et al. in a retrospective study, showed sensitivities and specificities around 80% [18]. However, this was only measured on tissue and not correlated to blood markers.
SALL4, a zinc-finger transcription factor, was evaluated in a prospective study by Khales et al. in 2015 and showed sensitivities of 96.1% and specificity of 95% [19]. This transcription factor is also found in other cancers and needs further validation in a larger cohort of patients which include polyps [19].
Cytokines (Table 4)
These are small secreted proteins, which can have autocrine or paracrine effects. Types of cytokines include chemokines (e.g., interleukin-8), lymphokines (e.g., interleukin-6), and interferons. They are released by a variety of cells including macrophages, T cells, B cells, and mast cells, and have been implicated in inflammatory and neoplastic diseases. The most promising was interleukin-8, which showed a sensitivity of 70% and specificity of 91% in detecting CRC in a meta-analysis conducted by Xia WJ et al. in 2015 [20]. Interleukin-8 is a chemokine thought to be involved in cancer progression and promotes angiogenesis, proliferation and migration of the cancer cells [20]. The study included 5 diagnostic studies with 725 participants. They were all high quality studies and if 1 study, by Burger et al., was excluded, the heterogeneity was significantly reduced. Limitations to this study were that it includes a relatively small selection of studies, the cutoffs of the different studies varied and a subgroup analysis could not be performed [20].
Antibodies (Table 5)
There were only three studies in the last 5 years that investigated antibodies, which fulfilled the inclusion criteria. None of the studies showed promising enough results for their use in diagnosis of CRC.
Proteins (Table 6)
In this category, the more promising proteins were trefoil factor (TFF)3 [21], CA11-19 [22], a combination of insulin-like growth factor binding protein 2 (IGFBP2), Dickkopf-3(DKK3), and pyruvate kinase M2 (PKM2) [23] and DC-SIGN/DC-SIGNR [24].
TFF3 belongs to a TFF family, which consists of three stable secretory proteins: TFF1-3. TFF3 is secreted by goblet cells of the intestine and to a lesser extent in the salivary glands, breast, and respiratory tissue. It is thought to promote invasion of cells by acting directly on the cells and indirectly on the vasculature. Results from the study by Li et al. showed a sensitivity of 74.2% and specificity of 94.8%; however, the level of this marker in polyps is not significantly different from that in the CRC cohort, making this less likely to be a useful diagnostic test [21].
CA11-19 is a 701 amino acid glycoprotein which showed very promising results in detecting CRC in a study by Overholt et al [22]. It showed a sensitivity of only 40% in detecting adenomatous polyps, but again a larger study is needed to include more CRC and more patients with polyps. Other limitations for this study were that only one center was included and the authors indicated a larger multi-center study was being planned [22].
Fung KY et al. investigated a combination of IGFBP2DKK3 and PKM2 [23] which have been implicated in proliferation, migration, and angiogenesis of cancer cells. The study showed a sensitivity of 73% and specificity of 95% in detecting CRC [23]. The sensitivity of detecting early stage cancer was moderate with 59% in stage I and 84% in stage II. This again needs further validation in a larger study, which includes patients with polyps.
DC-SIGN and DC-SIGNR are membrane-bound C type lectins. DC-SIGN is found on the surface of dendritic cells in the colon but also in the placenta, cervical mucosa and uterus. DC-SIGNR is found on the endothelial cells in the placenta, liver, and lymph nodes. Jian YM et al. investigated the serum level of DC-SIGN and DC-SIGNR, showing very high sensitivity and specificity in detecting CRC from healthy controls in a 290-patient cohort. The markers (DC-SIGN and DC-SIGNR) were separately analyzed for their sensitivity of detecting early CRC (stage I–III) and this was higher compared to CEA/CA19-9, yet polyps were not investigated [24].
Selection of markers for outcome study
From our review the following markers were found to have a sensitivity ≥ 70% and specificity ≥ 90%: interleukin 8 [25], NEAT_v2, SDC2 methylation of DNA [17], SEPT9 [14, 26], CA11-19 [22] and DC-SIGN/DC-SIGNR [24]. This cutoff sensitivity and specificity were used because they will discriminate markers which show promise in detecting CRC, and are comparable to the currently available biomarkers. The primary, secondary, and tertiary outcomes for these markers are as follows.
Diagnostic ability summary
Interleukin 8 (sensitivity 70%, specificity 91%, AUC 0.92), NEAT_v2 (sensitivity 70%, specificity 96%, AUC 0.871), SDC2 methylation of DNA (sensitivity 87%, specificity 95.2%), SEPT9 (sensitivity 71%, specificity 92%), CA11-19 (sensitivity 98%, specificity 84%), and DC-SIGN/DC-SIGNR (sensitivity 94.8%, specificity 98.7%, AUC 0.9885).
Comparison studies summary
The DC-SIGN/DC-SIGNR was compared to CEA (sensitivity 29.22%) and CA 19-9 (sensitivity 14.67%) and had better sensitivity (98.7%) and specificity (94.8%). SEPT9 in combination with fecal immunochemical test (FIT) improved sensitivity to 94% from 71% but decreased specificity to 68% from 92% in a recent meta-analysis by Nian et al [14]. Compared to FIT alone, Jin and colleagues showed that septin 9 showed a sensitivity and specificity of detecting CRC of 74.8 and 87.4%, respectively, whereas FIT alone had 58% sensitivity and 82.4% specificity [26]. CA 11-19, SDC2 methylation of DNA, NEAT1_v1 and Il-8 were not compared to any other marker.
Early stage detection summary
The tertiary outcome was assessed in SDC-SIGN/SDC-SIGNR, SEPT9, and SDC2 methylation of DNA and NEAT1_v1. NEAT1_v1 showed no difference in the mean value of the marker in early compared to late cancers. For SEPT9, sensitivity of detecting stage I CRC was 45% and 70% for stage II [14]. SDC2 methylation of DNA showed a sensitivity of 92% for detecting stage I CRC [17]. Finally, SDC-SIGN showed sensitivity and specificity of detecting early CRC of 81.33% and 55.56%, respectively [24].
Discussion
Investigation of biomarkers for the diagnosis of CRC can have a significant effect on its prognosis. Ransohoff described the search for a non-invasive biomarker as the “Holy Grail of cancer biomarker research” [27].
The currently used screening tests are either too uncomfortable, costly and potentially hazardous or have a low compliance rate due to patients’ aversion to sampling stool.
This review has highlighted the large numbers of markers being investigated and yet there is a lack of well-designed studies to investigate their use in diagnosis of CRC. There is also a lack of follow-up studies for many markers which have shown promise.
One of the first blood tests brought to clinical use for screening is called ColonSentry™. This measures mRNA in a 7-gene panel (ANXA3, CLEC4D, LMNB1, PRRG4, TNFAIP6, VNN1, and IL2RB) [27]. The sensitivity and specificity for ColonSentry for detecting CRC is 72 and 70%, respectively [27]. Retrospective studies on methylated SEPT9, have reported sensitivities, which range from 52 to 72% and specificities which range from 90 to 95% in detecting CRC [28, 29]. However, methylated SEPT9 has also been studied in a screening population as part of the PRESEPT trial in the USA and Germany, showing overall sensitivity of 48.2% and specificity of 91.5% for detecting CRC [30]. The difference between the PRESEPT and the other studies was that the first investigated the screening capacity of asymptomatic patients rather than symptomatic ones.
The markers we identified as promising included the following: interleukin 8 [25], NEAT_v2, SDC2 methylation of DNA [17], SEPT9 [14, 26], CA11-19 [22] and DC-SIGN/DC-SIGNR [24]. However, none were ideal and many had limitations that need addressing. Meta-analysis had been performed for only two markers (IL-8 and SEPT9) and the first included a small number of studies with variable cutoff values and a subgroup analysis could not be performed. As for the meta-analysis for SEPT9 published by Nian et al. in 2017, only two studies showed a low risk of bias. A total of 25 studies were included and most of them used the 2/3 positive result (known as the 2/3 algorithm) of the Epipro Colon assay. The pooled sensitivity was 71% and specificity 92% for generation 1 Epipro Colon assay and 76% and 94%, respectively, for generation 2 assay. The diagnostic value was highest for stage IV disease with a sensitivity of 79% specificity of 93%. However, 12 of the studies showed that SEPT9 has a sensitivity of 15 and 5% in detecting adenomas and polyps, respectively, and pooled sensitivity for larger size (> 1 cm) polyps or adenomas was 23%, making it a less than ideal screening marker. The studies which combined FIT with SEPT9 showed a higher sensitivity (94%) but lower specificity (68%). A recent retrospective study by Fu and colleagues also found that the 1/3 positive results (known as the 1/3 algorithm) of Epipro Colon Assay 2.0, is more sensitive in detecting early CRC (sensitivity of 69.6% for stage I) although still poor at detecting polyps and adenomas (sensitivity 16.8% from 7.9% using the 2/3 algorithm) and therefore, may be useful as an early cancer screening test [31]. Although SEPT9 shows promise, there is a large heterogeneity in the study results, which may be attributed to many factors including gender, race, age, assay method, and other environmental factors so larger prospective studies are required to further verify its diagnostic potential. SDC-SIGN, SDC-SIGNR, SEPT 9, and SDC2 methylation of DNA also showed a better detection rate in early versus late CRC compared to CEA. However, the sensitivities and specificities were still too low to have any true value in diagnosing CRC.
The rest of the studies did not clearly define their control groups, many including polyps in the control group and others not including them at all. Patient cohorts were mixed between symptomatic and asymptomatic individuals, which affected the results. Furthermore, not all studies assessed their markers’ ability to detect early CRC, an important factor in a diagnostic/screening marker. Variability of cutoff values, method of analysis of the markers, and timing of sample taking can also have a significant impact on the heterogeneity of the results. Inclusion of positive controls is also important as many of these markers, test positive in other cancers or diseases.
The cost-effectiveness of the test is another important consideration. In 2009, Lansdorp-Vogelaar and colleagues concluded that investigations such as endoscopy, FOBT, and FIT were all cost-effective given the high cost of treating late-stage CRC [32]. Ladabaum and colleagues who investigated the cost-effectiveness of SEPT9 as a screening test in Germany found that although it is more cost-effective than no screening at all, it is less cost-effective than FIT [33]. It is important to note that this model was based on a prospective study on asymptomatic patients, which showed lower sensitivity as we have discussed above. These authors have not found any studies investigating the cost-effectiveness of all the other markers identified in this study as promising in diagnosis of CRC. It is important to consider the combination of biomarkers or even combination of blood test and stool-based tests, to increase the accuracy of the test.
Among the limitations of this review is the exclusion of studies that looked at diagnostic markers for patients with adenomas or polyps but no CRC. Moreover, not all studies included all relevant demographic details on patients, whether the tumors were colonic (right versus left-sided) or rectal which can underestimate the diagnostic potential of the markers. Even with studies assessing the same marker, the assay method and cutoff values were not always homogeneous, thus introducing more variability in the results.
Conclusions
The race is still on to discover a sensitive, specific blood-based test for the diagnosis of CRC.
References
UK CR (2018) Bowel cancer mortality statistics. http://www.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/bowel-cancer/mortality. Accessed 1 Apr 2018
Heiss JA, Brenner H (2017) Epigenome-wide discovery and evaluation of leukocyte DNA methylation markers for the detection of colorectal cancer in a screening setting. Clin Epigenetics 9:24. https://doi.org/10.1186/s13148-017-0322-x
Toiyama Y, Tanaka K, Inoue Y, Mohri Y, Kusunoki M (2016) Circulating cell-free microRNAs as biomarkers for colorectal cancer. Surg Today 46(1):13–24. https://doi.org/10.1007/s00595-015-1138-y
Lin J, Chuang C-C, Zuo L (2017) Potential roles of microRNAs and ROS in colorectal cancer: diagnostic biomarkers and therapeutic targets. Oncotarget. https://doi.org/10.18632/oncotarget.14461
Young GP, Pedersen SK, Mansfield S, Murray DH, Baker RT, Rabbitt P, Byrne S, Bambacas L, Hollington P, Symonds EL (2016) A cross-sectional study comparing a blood test for methylated BCAT1 and IKZF1 tumor-derived DNA with CEA for detection of recurrent colorectal cancer. Cancer Med 5(10):2763–2772. https://doi.org/10.1002/cam4.868
Vatandoost N, Ghanbari J, Mojaver M, Avan A, Ghayour-Mobarhan M, Nedaeinia R, Salehi R (2016) Early detection of colorectal cancer: from conventional methods to novel biomarkers. J Cancer Res Clin Oncol 142(2):341–351. https://doi.org/10.1007/s00432-015-1928-z
Brenner H, Stock C, Hoffmeister M (2015) Colorectal cancer screening: the time to act is now. BMC Med. https://doi.org/10.1186/s12916-015-0498-x
Lim DH, Lee JH, Kim JW (2017) Feasibility of CYFRA 21—1 as a serum biomarker for the detection of colorectal adenoma and advanced colorectal adenoma in people over the age of 45. J Clin Lab Anal. https://doi.org/10.1002/jcla.22163
Werner S, Krause F, Rolny V, Strobl M, Morgenstern D, Datz C, Chen H, Brenner H (2016) Evaluation of a 5-marker blood test for colorectal cancer early detection in a colorectal cancer screening setting. Clin Cancer Res 22(7):1725–1733. https://doi.org/10.1158/1078-0432.ccr-15-1268
New bowel cancer screening test (2016). https://www.gov.uk/government/news/new-bowel-cancer-screening-test. Accessed 13 Dec 2017
Imaoka H, Toiyama Y, Fujikawa H, Hiro J, Saigusa S, Tanaka K, Inoue Y, Mohri Y, Mori T, Kato T, Toden S, Goel A, Kusunoki M (2016) Circulating microRNA-1290 as a novel diagnostic and prognostic biomarker in human colorectal cancer. Ann Oncol 27(10):1879–1886. https://doi.org/10.1093/annonc/mdw279
Zhang HH, Li PW, Ju HX, Pesta M, Kulda V, Jin WJ, Cai M, Liu CB, Wu H, Xu JM, Ye Y, Zhang GL, Xu EP, Cai JT, Lai MD, Xia DJ, Yang J, Wu YH (2014) Diagnostic and prognostic value of microRNA-21 in colorectal cancer: an original study and individual participant data meta-analysis. Cancer Epidemiol Biomark Prev 23(12):2783–2792. https://doi.org/10.1158/1055-9965.epi-14-0598
Fang Z, Tang J, Bai Y, Lin H, You H, Jin H, Lin L, You P, Li J, Dai Z, Liang X, Su Y, Hu Q, Wang F, Zhang Z-Y (2015) Plasma levels of microRNA-24, microRNA-320a, and microRNA-423-5p are potential biomarkers for colorectal carcinoma. J Exp Clin Cancer Res 34:86. https://doi.org/10.1186/s13046-015-0198-6
Nian J, Sun X, Ming S, Yan C, Ma Y, Feng Y, Yang L, Yu M, Zhang G, Wang X (2017) Diagnostic accuracy of methylated SEPT9 for blood-based colorectal cancer detection: a systematic review and meta-analysis. Clin Transl Gastroenterol 8(1):e216. https://doi.org/10.1038/ctg.2016.66
Wu YC, Yang L, Zhao J, Li C, Nie J, Liu FQ, Zhuo CH, Zheng YX, Li B, Wang ZM, Xu Y (2015) Nuclear-enriched abundant transcript 1 as a diagnostic and prognostic biomarker in colorectal cancer. Mol Cancer. https://doi.org/10.1186/s12943-015-0455-5
Hao TB, Shi W, Shen XJ, Qi J, Wu XH, Wu Y, Tang YY, Ju SQ (2014) Circulating cell-free DNA in serum as a biomarker for diagnosis and prognostic prediction of colorectal cancer. Br J Cancer 111(8):1482–1489. https://doi.org/10.1038/bjc.2014.470
Oh T, Kim N, Moon Y, Kim MS, Hoehn BD, Park CH, Kim TS, Kim NK, Chung HC, An S (2013) Genome-wide identification and validation of a novel methylation biomarker, SDC2, for blood-based detection of colorectal cancer. J Mol Diagn 15(4):498–507. https://doi.org/10.1016/j.jmoldx.2013.03.004
Li W, Zhang G, Wang HL, Wang L (2016) Analysis of expression of cyclin E, p27kip1 and Ki67 protein in colorectal cancer tissues and its value for diagnosis, treatment and prognosis of disease. Eur Rev Med Pharmacol Sci 20(23):4874–4879
Khales SA, Abbaszadegan MR, Abdollahi A, Raeisossadati R, Tousi MF, Forghanifard MM (2015) SALL4 as a new biomarker for early colorectal cancers. Cancer Res Clin Oncol 141(2):229–235. https://doi.org/10.1007/s00432-014-1808-y
Xia WJ, Chen WZ, Zhang ZG, Wu D, Wu P, Chen ZG, Li C, Huang J (2015) Prognostic value, clinicopathologic features and diagnostic accuracy of interleukin-8 in colorectal cancer: a meta-analysis. PloS One. https://doi.org/10.1371/journal.pone.0123484
Li Q, Wang K, Su C, Fang J (2016) Serum trefoil factor 3 as a protein biomarker for the diagnosis of colorectal cancer. Technol Cancer Res Treat. https://doi.org/10.1177/1533034616674323
Overholt BF, Wheeler DJ, Jordan T, Fritsche HA (2016) CA11-19: a tumor marker for the detection of colorectal cancer. Gastrointest Endosc 83(3):545–551. https://doi.org/10.1016/j.gie.2015.06.041
Fung KYC, Tabor B, Buckley MJ, Priebe IK, Purins L, Pompeia C, Brierley GV, Lockett T, Gibbs P, Tie J, McMurrick P, Moore J, Ruszkiewicz A, Nice E, Adams TE, Burgess A, Cosgrove LJ (2015) Blood-based protein biomarker panel for the detection of colorectal cancer. PLoS One. https://doi.org/10.1371/journal.pone.0120425
Jiang Y, Zhang C, Chen K, Chen Z, Sun Z, Zhang Z, Ding D, Ren S, Zuo Y (2014) The clinical significance of DC-SIGN and DC-SIGNR, which are novel markers expressed in human colon cancer. PLoS One 9(12):e114748. https://doi.org/10.1371/journal.pone.0114748
Jin WJ, Xu JM, Xu WL, Gu DH, Li PW (2014) Diagnostic value of interleukin-8 in colorectal cancer: a case–control study and meta-analysis. World J Gastroenterol 20(43):16334–16342. https://doi.org/10.3748/wjg.v20.i43.16334
Jin P, Kang Q, Wang X, Yang L, Yu Y, Li N, He YQ, Han XL, Hang J, Zhang J, Song LL, Han Y, Sheng JQ (2015) Performance of a second-generation methylated SEPT9 test in detecting colorectal neoplasm. J Gastroenterol Hepatol 30(5):830–833. https://doi.org/10.1111/jgh.12855
Heichman KA (2014) Blood-based testing for colorectal cancer screening. Mol Diagn Ther 18(2):127–135. https://doi.org/10.1007/s40291-013-0074-z
Lofton-Day C, Model F, Devos T, Tetzner R, Distler J, Schuster M, Song X, Lesche R, Liebenberg V, Ebert M, Molnar B, Grutzmann R, Pilarsky C, Sledziewski A (2008) DNA methylation biomarkers for blood-based colorectal cancer screening. Clin Chem 54(2):414–423. https://doi.org/10.1373/clinchem.2007.095992
deVos T, Tetzner R, Model F, Weiss G, Schuster M, Distler J, Steiger KV, Grutzmann R, Pilarsky C, Habermann JK, Fleshner PR, Oubre BM, Day R, Sledziewski AZ, Lofton-Day C (2009) Circulating methylated SEPT9 DNA in plasma is a biomarker for colorectal cancer. Clin Chem 55(7):1337–1346. https://doi.org/10.1373/clinchem.2008.115808
Church T (2014) Colorectal cancer screening: will non-invasive procedures triumph? Genome Med. https://doi.org/10.1186/gm562
Fu B, Yan P, Zhang S, Lu Y, Pan L, Tang W, Chen S, Chen S, Zhang A, Liu W (2018) Cell-free circulating methylated SEPT9 for noninvasive diagnosis and monitoring of colorectal cancer. Dis Mark 2018:6437104. https://doi.org/10.1155/2018/6437104
Lansdorp-Vogelaar I, van Ballegooijen M, Zauber AG, Habbema JD, Kuipers EJ (2009) Effect of rising chemotherapy costs on the cost savings of colorectal cancer screening. J Natl Cancer Inst 101(20):1412–1422. https://doi.org/10.1093/jnci/djp319
Ladabaum U, Alvarez-Osorio L, Rösch T, Brueggenjuergen B (2014) Cost-effectiveness of colorectal cancer screening in Germany: current endoscopic and fecal testing strategies versus plasma methylated Septin 9 DNA. Endosc Int Open 2(2):E96–E104. https://doi.org/10.1055/s-0034-1377182
Li L, Guo Y, Chen Y, Wang J, Zhen L, Guo X, Liu J, Jing C (2016) The diagnostic efficacy and biological effects of microRNA-29b for colon cancer. Technol Cancer Res Treat 15(6):772–779
Basati G, Razavi AE, Pakzad I, Malayeri FA (2016) Circulating levels of the miRNAs, miR-194, and miR-29b, as clinically useful biomarkers for colorectal cancer. Tumour Biol J Int Soc Oncodevelopmental Biol Med 37(2):1781–1788. https://doi.org/10.1007/s13277-015-3967-0
Zhi ML, Liu ZJ, Yi XY, Zhang LJ, Bao YX (2015) Diagnostic performance of microRNA-29a for colorectal cancer: a meta-analysis. Genet Mol Res 14(4):18018–18025. https://doi.org/10.4238/2015.December.22.28
Xu F, Xu L, Wang M, An G, Feng G (2015) The accuracy of circulating microRNA-21 in the diagnosis of colorectal cancer: a systematic review and meta-analysis. Colorectal Dis 17(5):O100–O107. https://doi.org/10.1111/codi.12917
Lv ZC, Fan YS, Chen HB, Zhao DW (2015) Investigation of microRNA-155 as a serum diagnostic and prognostic biomarker for colorectal cancer. Tumour Biol J Int Soc Oncodevelopmental Biol Med 36(3):1619–1625. https://doi.org/10.1007/s13277-014-2760-9
Chen WY, Zhao XJ, Yu ZF, Hu FL, Liu YP, Cui BB, Dong XS, Zhao YS (2015) The potential of plasma miRNAs for diagnosis and risk estimation of colorectal cancer. Int J Clin Exp Pathol 8(6):7092–7101
Zhang J, Zhang KJ, Bi MS, Jiao XL, Zhang DL, Dong Q (2014) Circulating microRNA expressions in colorectal cancer as predictors of response to chemotherapy. Anti Cancer Drugs 25(3):346–352. https://doi.org/10.1097/cad.0000000000000049
Yang X, Zeng ZY, Hou YX, Yuan TX, Gao C, Jia W, Yi XY, Liu MR (2014) MicroRNA-92a as a potential biomarker in diagnosis of colorectal cancer: a systematic review and meta-analysis. PLoS One. https://doi.org/10.1371/journal.pone.0088745
Xu LL, Li MZ, Wang M, Yan D, Feng GS, An GY (2014) The expression of microRNA-375 in plasma and tissue is matched in human colorectal cancer. BMC Cancer. https://doi.org/10.1186/1471-2407-14-714
Nonaka R, Nishimura J, Kagawa Y, Osawa H, Hasegawa J, Murata K, Okamura S, Ota H, Uemura M, Hata T, Takemasa I, Mizushima T, Okuzaki D, Yamamoto H, Doki Y, Mori M (2014) Circulating miR-199a-3p as a novel serum biomarker for colorectal cancer. Oncol Rep 32(6):2354–2358. https://doi.org/10.3892/or.2014.3515
Zhu M, Huang Z, Zhu D, Zhou X, Shan X, Qi LW, Wu L, Cheng W, Zhu J, Zhang L, Zhang H, Chen Y, Zhu W, Wang T, Liu P (2017) A panel of microRNA signature in serum for colorectal cancer diagnosis. Oncotarget. https://doi.org/10.18632/oncotarget.15059
Vychytilova-Faltejskova P, Radova L, Sachlova M, Kosarova Z, Slaba K, Fabian P, Grolich T, Prochazka V, Kala Z, Svoboda M, Kiss I, Vyzula R, Slaby O (2016) Serum-based microRNA signatures in early diagnosis and prognosis prediction of colon cancer. Carcinogenesis 37(10):941–950. https://doi.org/10.1093/carcin/bgw078
Fang ZX, Tang J, Bai YY, Lin HY, You HY, Jin HW, Lin LQ, You P, Li J, Dai Z, Liang XM, Su YH, Hu Q, Wang F, Zhang ZY (2015) Plasma levels of microRNA-24, microRNA-320a, and microRNA-423-5p are potential biomarkers for colorectal carcinoma. J Exp Clin Cancer Res. https://doi.org/10.1186/s13046-015-0198-6
Wang J, Huang SK, Zhao M, Yang M, Zhong JL, Gu YY, Peng H, Che YQ, Huang CZ (2014) Identification of a circulating microRNA signature for colorectal cancer detection. PLoS One 9(4):e87451. https://doi.org/10.1371/journal.pone.0087451
Zhang GJ, Zhou T, Liu ZL, Tian HP, Xia SS (2013) Plasma miR-200c and miR-18a as potential biomarkers for the detection of colorectal carcinoma. Mol Clin Oncol 1(2):379–384. https://doi.org/10.3892/mco.2013.61
Luo X, Stock C, Burwinkel B, Brenner H (2013) Identification and evaluation of plasma microRNAs for early detection of colorectal cancer. PLoS One 8(5):e62880. https://doi.org/10.1371/journal.pone.0062880
Nagai Y, Sunami E, Yamamoto Y, Hata K, Okada S, Murono K, Yasuda K, Otani K, Nishikawa T, Tanaka T, Kiyomatsu T, Kawai K, Nozawa H, Ishihara S, Hoon DSB, Watanabe T (2017) LINE-1 hypomethylation status of circulating cell-free DNA in plasma as a biomarker for colorectal cancer. Oncotarget. https://doi.org/10.18632/oncotarget.14439
Pedersen SK, Symonds EL, Baker RT, Murray DH, McEvoy A, Van Doorn SC, Mundt MW, Cole SR, Gopalsamy G, Mangira D, LaPointe LC, Dekker E, Young GP (2015) Evaluation of an assay for methylated BCAT1 and IKZF1 in plasma for detection of colorectal neoplasia. BMC Cancer 15:654. https://doi.org/10.1186/s12885-015-1674-2
Qi J, Qian C, Shi W, Wu XH, Jing RR, Zhang LR, Wang ZW, Ju SQ (2013) Alu-based cell-free DNA: a potential complementary biomarker for diagnosis of colorectal cancer. Clin Biochem 46(1–2):64–69. https://doi.org/10.1016/j.clinbiochem.2012.08.026
Rodia MT, Ugolini G, Mattei G, Montroni I, Zattoni D, Ghignone F, Veronese G, Marisi G, Lauriola M, Strippoli P, Solmi R (2016) Systematic large-scale meta-analysis identifies a panel of two mRNAs as blood biomarkers for colorectal cancer detection. Oncotarget 7(21):30295–30306. https://doi.org/10.18632/oncotarget.8108
Wang X, Yang Z, Tian H, Li Y, Li M, Zhao W, Zhang C, Wang T, Liu J, Zhang A, Shen D, Zheng C, Qi J, Zhao D, Shi J, Jin L, Rao J, Zhang W (2017) Circulating MIC-1/GDF15 is a complementary screening biomarker with CEA and correlates with liver metastasis and poor survival in colorectal cancer. Oncotarget. https://doi.org/10.18632/oncotarget.15279
Xu J, Ye Y, Zhang H, Szmitkowski M, Makinen MJ, Li P, Xia D, Yang J, Wu Y, Wu H (2016) Diagnostic and Prognostic value of serum interleukin-6 in colorectal cancer. Medicine 95(2):e2502. https://doi.org/10.1097/md.0000000000002502
Zheng ZX, Zheng M, Bi JJ, Feng Q, Yue ZG, Zhou YQ, Hu WN, Zhang HZ, Gao HJ (2015) Serum GRO beta: a potential tumor-associated biomarker for colorectal cancer. Int J Clin Exp Med 8(2):2526–2535
Kunizaki M, Sawai T, Takeshita H, Tominaga T, Hidaka S, To K, Miyazaki T, Hamamoto R, Nanashima A, Nagayasu T (2016) Clinical value of serum p53 antibody in the diagnosis and prognosis of colorectal cancer. Anticancer Res 36(8):4171–4175
Chen H, Werner S, Butt J, Zornig I, Knebel P, Michel A, Eichmuller SB, Jager D, Waterboer T, Pawlita M, Brenner H (2016) Prospective evaluation of 64 serum autoantibodies as biomarkers for early detection of colorectal cancer in a true screening setting. Oncotarget 7(13):16420–16432. https://doi.org/10.18632/oncotarget.7500
Wang HF, Li LF, Guo SH, Zeng QY, Ning F, Liu WL, Zhang G (2016) Evaluation of antibody level against Fusobacterium nucleatum in the serological diagnosis of colorectal cancer. Sci Rep 6:33440. https://doi.org/10.1038/srep33440
Fei W, Chen L, Chen J, Shi Q, Zhang L, Liu S, Li L, Zheng L, Hu X (2017) RBP4 and THBS2 are serum biomarkers for diagnosis of colorectal cancer. Oncotarget 8(54):92254–92264. https://doi.org/10.18632/oncotarget.21173
Wang X-Q, Tang Z-X, Yu D, Cui S-J, Jiang Y-H, Zhang Q, Wang J, Yang P-Y, Liu F (2016) Epithelial but not stromal expression of collagen alpha-1(III) is a diagnostic and prognostic indicator of colorectal carcinoma. Oncotarget 7(8):8823–8838. https://doi.org/10.18632/oncotarget.6815
Gezer U, Yoruker EE, Keskin M, Kulle CB, Dharuman Y, Holdenrieder S (2015) Histone methylation marks on circulating nucleosomes as novel blood-based biomarker in colorectal cancer. Int J Mol Sci 16(12):29654–29662. https://doi.org/10.3390/ijms161226180
Xue YM, Yu FD, Yan DW, Cui FF, Tang HM, Wang XL, Chen J, Lu HJ, Zhao SL, Peng ZH (2015) Zinc-alpha-2-glycoprotein: a candidate biomarker for colon cancer diagnosis in Chinese population. Int J Mol Sci 16(1):691–703. https://doi.org/10.3390/ijms16010691
Wang ZQ, Sun XL, Su HL, Liu XF, Xuan YJ, Yu SQ (2015) Association between serum angiopoietin-2 concentration and clinicopathological parameters in patients with colorectal cancer. Genet Mol Res 14(4):15547–15552. https://doi.org/10.4238/2015.December.1.5
Storm L, Christensen IJ, Jensenius JC, Nielsen HJ, Thiel S, Danish Study Grp Early D (2015) Evaluation of complement proteins as screening markers for colorectal cancer. Cancer Immunol Immunother 64(1):41–50. https://doi.org/10.1007/s00262-014-1615-y
Fung KY, Tabor B, Buckley MJ, Priebe IK, Purins L, Pompeia C, Brierley GV, Lockett T, Gibbs P, Tie J, McMurrick P, Moore J, Ruszkiewicz A, Nice E, Adams TE, Burgess A, Cosgrove LJ (2015) Blood-based protein biomarker panel for the detection of colorectal cancer. PLoS One 10(3):e0120425. https://doi.org/10.1371/journal.pone.0120425
Sole X, Crous-Bou M, Cordero D, Olivares D, Guino E, Sanz-Pamplona R, Rodriguez-Moranta F, Sanjuan X, de Oca J, Salazar R, Moreno V (2014) Discovery and validation of new potential biomarkers for early detection of colon cancer. PLoS One 9(9):e106748. https://doi.org/10.1371/journal.pone.0106748
Shin J, Kim HJ, Kim G, Song M, Woo SJ, Lee ST, Kim H, Lee C (2014) Discovery of melanotransferrin as a serological marker of colorectal cancer by secretome analysis and quantitative proteomics. J Proteome Res 13(11):4919–4931. https://doi.org/10.1021/pr500790f
Shirahata A, Hibi K (2014) Serum vimentin methylation as a potential marker for colorectal cancer. Anticancer Res 34(8):4121–4125
Wang J, Wang X, Lin S, Chen C, Wang C, Ma Q, Jiang B (2013) Identification of kininogen-1 as a serum biomarker for the early detection of advanced colorectal adenoma and colorectal cancer. PLoS One 8(7):e70519. https://doi.org/10.1371/journal.pone.0070519
Funding
There was no funding source for this study.
Author information
Authors and Affiliations
Contributions
All the authors have contributed significantly and are in agreement with the content of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare they have no conflict of interest.
Ethical approval
This article is compliant with ethical standards.
Informed consent
Informed consent was not required for this study.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Nikolaou, S., Qiu, S., Fiorentino, F. et al. Systematic review of blood diagnostic markers in colorectal cancer. Tech Coloproctol 22, 481–498 (2018). https://doi.org/10.1007/s10151-018-1820-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10151-018-1820-3