Abstract
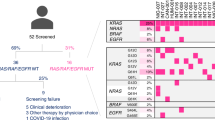
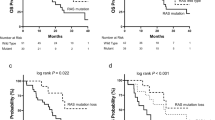
Oncogenic RAS mutations are negative biomarkers of response to epidermal growth factor receptor (EGFR) blockade. RAS mutations are usually detected in biopsies of primary colorectal tumors. However, the genomic profiles of primary tumors and metastases are not always concordant, and chemotherapeutic agents can alter the tumor molecular landscape. Cell-free DNA (cfDNA) is a novel tool to detect molecular heterogeneity. This study evaluated the clinical utility of cfDNA to predict primary or secondary resistance to EGFR blockade in patients with metastatic colorectal cancer. Thirty metastatic colorectal cancer patients without RAS and BRAF mutations were prospectively enrolled and treated with cytotoxic agents and EGFR blockade as first-line therapy. cfDNA was analyzed for the presence of RAS, BRAF, and EGFR (S492R) point mutations before initiating chemotherapy and every 2 months during chemotherapy. The analysis was performed in 223 plasma samples from all 30 patients. Of the 30 patients, five had RAS mutations in their cfDNA before starting chemotherapy and did not respond. Twenty-four of the remaining 25 patients without cfDNA RAS mutations had a response. Twenty of the 24 responders developed secondary resistance and cfDNA RAS mutations were found in 17 of the 20. cfDNA BRAF mutations were found in seven, and EGFR mutations were found in eight of the 20 patients. Emerging RAS, BRAF, and EGFR mutations occurred in patients with primary and secondary resistance to EGFR blockade. The detection of these mutations in cfDNA is a promising approach to predict treatment response and secondary resistance.



Similar content being viewed by others
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- EGFR:
-
Epidermal growth factor receptor
- cfDNA:
-
Cell-free DNA
- mCRC:
-
Metastatic colorectal cancer
- ETS:
-
Early tumor shrinkage
- CR:
-
Complete response
- PR:
-
Partial response
- SD:
-
Stable disease
- RECIST:
-
Response Evaluation Criteria in Solid Tumors
- dPCR:
-
Digital PCR
References
Cunningham D, Humblet Y, Siena S et al (2004) Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Eng J Med 351:337–345
Piessevaux H, Buyse M, Schlichting M et al (2013) Use of early tumor shrinkage to predict long-term outcome in metastatic colorectal cancer treated with cetuximab. J Clin Oncol 31:3764–3775
Matsuhashi N, Takahashi T, Matsui S et al (2018) A novel therapeutic strategy of personalized medicine based on anti-epidermal growth factor receptor monoclonal antibodies in patients with metastatic colorectal cancer. Int J Oncol 52:1391–1400
Lievre A, Bachet JB, Le Corre D et al (2006) KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Can Res 66:3992–3995
Douillard JY, Oliner KS, Siena S et al (2013) Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Eng J Med 369:1023–1034
Baldus SE, Schaefer KL, Engers R et al (2010) Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin Cancer Res 16:790–799
Mao C, Wu XY, Yang ZY et al (2015) Concordant analysis of KRAS, BRAF, PIK3CA mutations, and PTEN expression between primary colorectal cancer and matched metastases. Sci Rep 5:8065
Vogelstein B, Papadopoulos N, Velculescu VE et al (2013) Cancer genome landscapes. Science (New York, N.Y.) 339:1546–1558
Yamada T, Matsuda A, Koizumi M et al (2019) Liquid biopsy for the management of patients with colorectal cancer. Digestion 99:39–45
Finotti A, Allegretti M, Gasparello J et al (2018) Liquid biopsy and PCR-free ultrasensitive detection systems in oncology (Review). Int J Oncol 53:1395–1434
Iwai T, Yamada T, Takahashi G et al (2019) Circulating cell-free long DNA fragments predict post-hepatectomy recurrence of colorectal liver metastases. Euro J Surg Oncol 46:108–114
Yamada T, Iwai T, Takahashi G et al (2016) Utility of KRAS mutation detection using circulating cell-free DNA from patients with colorectal cancer. Cancer Sci 107:936–943
Furuki H, Yamada T, Takahashi G et al (2018) Evaluation of liquid biopsies for detection of emerging mutated genes in metastatic colorectal cancer. Eur J Surg Oncol 44:975–982
Takeda K, Yamada T, Takahashi G et al (2019) Analysis of colorectal cancer-related mutations by liquid biopsy: utility of circulating cell-free DNA and circulating tumor cells. Cancer Sci 110:3497–3509
Vidal J, Muinelo L, Dalmases A et al (2017) Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann Oncol 28:1325–1332
Spindler KL, Pallisgaard N, Andersen RF et al (2014) Changes in mutational status during third-line treatment for metastatic colorectal cancer–results of consecutive measurement of cell free DNA, KRAS and BRAF in the plasma. Int J Cancer 135:2215–2222
Morelli MP, Overman MJ, Dasari A et al (2015) Characterizing the patterns of clonal selection in circulating tumor DNA from patients with colorectal cancer refractory to anti-EGFR treatment. Ann Oncol 26:731–736
Grasselli J, Elez E, Caratù G et al (2017) Concordance of blood- and tumor-based detection of RAS mutations to guide anti-EGFR therapy in metastatic colorectal cancer. Ann Oncol 28:1294–1301
Kim TW, Peeters M, Thomas A et al (2018) Impact of emergent circulating tumor dna ras mutation in panitumumab-treated chemoresistant metastatic colorectal cancer. Clin Cancer Res 24:5602–5609
Misale S, Yaeger R, Hobor S et al (2012) Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature 486:532–536
Diaz LA Jr, Williams RT, Wu J et al (2012) The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature 486:537–540
Siravegna G, Mussolin B, Buscarino M et al (2015) Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med 21:795–801
Yoshino T, Muro K, Yamaguchi K et al (2015) Clinical validation of a multiplex kit for RAS mutations in colorectal cancer: results of the RASKET (RAS KEy Testing) prospective. Multicenter Study EBioMedicine 2:317–323
Takahashi G, Yamada T, Iwai T et al (2018) Oncological assessment of stent placement for obstructive colorectal cancer from circulating cell-free DNA and circulating tumor DNA dynamics. Ann Surg Oncol 25:737–744
Thierry AR, Mouliere F, El Messaoudi S et al (2014) Clinical validation of the detection of KRAS and BRAF mutations from circulating tumor DNA. Nat Med 20:430–435
Spindler KL, Pallisgaard N, Vogelius I et al (2012) Quantitative cell-free DNA, KRAS, and BRAF mutations in plasma from patients with metastatic colorectal cancer during treatment with cetuximab and irinotecan. Clin Cancer Res 18:1177–1185
Taly V, Pekin D, Benhaim L et al (2013) Multiplex picodroplet digital PCR to detect KRAS mutations in circulating DNA from the plasma of colorectal cancer patients. Clin Chem 59:1722–1731
Siena S, Sartore-Bianchi A, Garcia-Carbonero R et al (2018) Dynamic molecular analysis and clinical correlates of tumor evolution within a phase II trial of panitumumab-based therapy in metastatic colorectal cancer. Ann Oncol 29:119–126
Pietrantonio F, Vernieri C, Siravegna G et al (2017) Heterogeneity of Acquired Resistance to Anti-EGFR Monoclonal Antibodies in Patients with Metastatic Colorectal Cancer. Clin Cancer Res 23:2414–2422
Xu J-M, Wang Y, Wang Y-L et al (2017) PIK3CA mutations contribute to acquired cetuximab resistance in patients with metastatic colorectal cancer. Clin Cancer Res 23:4602–4616
Thierry AR, Pastor B, Jiang Z-Q et al (2017) Circulating DNA demonstrates convergent evolution and common resistance mechanisms during treatment of colorectal cancer. Clin Cancer Res 23:4578–4591
Santini D, Loupakis F, Vincenzi B et al (2008) High concordance of KRAS status between primary colorectal tumors and related metastatic sites: implications for clinical practice. Oncologist 13:1270–1275
Gerlinger M, Rowan AJ, Horswell S et al (2012) Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Eng J Med 366:883–892
Yaeger R, Chatila WK, Lipsyc MD et al (2018) Clinical sequencing defines the genomic landscape of metastatic colorectal cancer. Cancer Cell 33:125–136
Van Emburgh BO, Arena S, Siravegna G et al (2016) Acquired RAS or EGFR mutations and duration of response to EGFR blockade in colorectal cancer. Nat Commun 7:13665
Raja R, Kuziora M, Brohawn P et al (2018) Early reduction in ctDNA predicts survival in lung and bladder cancer patients treated with durvalumab. Clin Cancer Res 24:6212–6222
Garlan F, Laurent-Puig P, Sefrioui D et al (2017) Early evaluation of circulating tumor DNA as marker of therapeutic efficacy in metastatic colorectal cancer patients (PLACOL Study). Clin Cancer Res 23:5416–5425
Yamauchi M, Urabe Y, Ono A et al (2017) Serial profiling of circulating tumor DNA for optimization of anti-VEGF chemotherapy in metastatic colorectal cancer patients. Int J Cancer 142:1418–1426
Acknowledgments
We thank Mr. Masaaki Higuchi for technical support in the experiments. We also thank H. Nikki March, PhD, from Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
Funding
This work was supported by JSPS KAKENHI Grant Number 17K10657. Funding was also provided locally by the Department of Gastrointestinal and Hepato-Biliary-Pancreatic Surgery, Nippon Medical School, Tokyo, Japan.
Author information
Authors and Affiliations
Contributions
TY designed this article, acquisitioned of the data and wrote this paper. MK, SS, GT, TI, KT, SK and KU acquisitioned, analyzed and interpreted the data. AM and RO checked the manuscript. HY supervised its quality and revised the paper. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
This study was approved by Medical Ethics Committee of Nippon Medical School, and written informed consent was obtained from the patients prior to sample collection. All experimental procedures were performed according to the regulations and internal biosafety and bioethics guidelines.
Patient consent for publication
Written informed consent was obtained from the patients.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
About this article
Cite this article
Yamada, T., Matsuda, A., Takahashi, G. et al. Emerging RAS, BRAF, and EGFR mutations in cell-free DNA of metastatic colorectal patients are associated with both primary and secondary resistance to first-line anti-EGFR therapy. Int J Clin Oncol 25, 1523–1532 (2020). https://doi.org/10.1007/s10147-020-01691-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10147-020-01691-0