Abstract
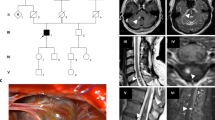
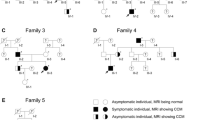
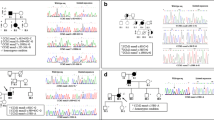
Familial cerebral cavernous malformations (CCMs) occur with a frequency of 1 in 2000 and may cause recurrent headaches, seizures, and hemorrhagic stroke. Exon-scanning-based methods have identified intragenic mutations in three genes, CCM1, CCM2, and CCM3, in about 70% of familial CCM. To date, only two large CCM2 and a single large CCM3 deletion have been published. In addition to direct sequencing of all three CCM genes, we applied a newly developed multiplex ligation-dependent probe amplification gene dosage assay (MLPA) designed to detect genomic CCM1–3 deletions/duplications. Direct sequencing did not reveal a mutation in the index case who presented with multiple CCMs that had caused a generalized tonic-clonic seizure with Todd’s paralysis and headaches at the age of 5. In contrast, MLPA analyses detected a large deletion involving the entire CCM1 coding region in the proband and further affected members of this German CCM family. The MLPA results were corroborated by analyses of single nucleotide polymorphisms (SNPs) within the CCM1 gene. Thus, we here present the first report on a CCM1 gene deletion. Our results confirm a loss-of-function mutation mechanism for CCM1 and demonstrate that the use of MLPA enables a higher CCM mutation detection rate which is crucial for predictive testing of at-risk relatives.



Similar content being viewed by others
References
Bergametti F, Denier C, Labauge P, Arnoult M, Boetto S, Clanet M, Coubes P, Echenne B, Ibrahim R, Irthum B, Jacquet G, Lonjon M, Moreau JJ, Neau JP, Parker F, Tremoulet M, Tournier-Lasserve E (2005) Mutations within the programmed cell death 10 gene cause cerebral cavernous malformations. Am J Hum Genet 76:42–51
Cavé-Riant F, Denier C, Labauge P, Cecillon M, Maciazek J, Joutel A, Laberge-Le Couteulx S, Tournier-Lasserve E (2002) Spectrum and expression analysis of KRIT1 mutations in 121 consecutive and unrelated patients with Cerebral Cavernous Malformations. Eur J Hum Genet 10:733–740
Damgaard D, Nissen PH, Jensen LG, Nielsen GG, Stenderup A, Larsen ML, Faergeman O (2005) Detection of large deletions in the LDL receptor gene with quantitative PCR methods. BMC Med Genet 6:15
Denier C, Goutagny S, Labauge P, Krivosic V, Arnoult M, Cousin A, Benabid AL, Comoy J, Frerebeau P, Gilbert B, Houtteville JP, Jan M, Lapierre F, Loiseau H, Menei P, Mercier P, Moreau JJ, Nivelon-Chevallier A, Parker F, Redondo AM, Scarabin JM, Tremoulet M, Zerah M, Maciazek J, Tournier-Lasserve E (2004) Mutations within the MGC4607 gene cause cerebral cavernous malformations. Am J Hum Genet 74:326–337
Guclu B, Ozturk AK, Pricola KL, Bilguvar K, Shin D, O’Roak BJ, Gunel M (2005) Mutations in apoptosis-related gene, PDCD10, cause cerebral cavernous malformation 3. Neurosurgery 57:1008–1013
Laberge-le Couteulx S, Jung HH, Labauge P, Houtteville JP, Lescoat C, Cecillon M, Marechal E, Joutel A, Bach JF, Tournier-Lasserve E (1999) Truncating mutations in CCM1, encoding KRIT1, cause hereditary cavernous angiomas. Nat Genet 23:189–193
Lalic T, Vossen RH, Coffa J, Schouten JP, Guc-Scekic M, Radivojevic D, Djurisic M, Breuning MH, White SJ, den Dunnen JT (2005) Deletion and duplication screening in the DMD gene using MLPA. Eur J Hum Genet 13:1231–1234
Liquori CL, Berg MJ, Siegel AM, Huang E, Zawistowski JS, Stoffer T, Verlaan D, Balogun F, Hughes L, Leedom TP, Plummer NW, Cannella M, Maglione V, Squitieri F, Johnson EW, Rouleau GA, Ptacek L, Marchuk DA (2003) Mutations in a gene encoding a novel protein containing a phosphotyrosine-binding domain cause type 2 cerebral cavernous malformations. Am J Hum Genet 73:1459–1464
Liquori CL, Berg MJ, Squitieri F, Ottenbacher M, Sorlie M, Leedom TP, Cannella M, Maglione V, Ptacek L, Johnson EW, Marchuk DA (2006) Low frequency of PDCD10 mutations in a panel of CCM3 probands: potential for a fourth CCM locus. Hum Mutat 27:118
Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G (2002) Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res 30:e57
Sürücü O, Sure U, Gaetzner S, Stahl S, Benes L, Bertalanffy H, Felbor U (2006) Clinical impact of CCM mutation detection in familial cavernous angioma. Childs Nerv Syst 22:1461–1464
Verlaan DJ, Roussel J, Laurent SB, Elger CE, Siegel AM, Rouleau GA (2005) CCM3 mutations are uncommon in cerebral cavernous malformations. Neurology 65:1982–1983
Verlaan DJ, Siegel AM, Rouleau GA (2002) Krit1 missense mutations lead to splicing errors in cerebral cavernous malformation. Am J Hum Genet 70:1564–1567
Wimmer K, Yao S, Claes K, Kehrer-Sawatzki H, Tinschert S, De Raedt T, Legius E, Callens T, Beiglbock H, Maertens O, Messiaen L (2006) Spectrum of single- and multiexon NF1 copy number changes in a cohort of 1,100 unselected NF1 patients. Genes Chromosomes Cancer 45:265–276
Zawistowski JS, Stalheim L, Uhlik MT, Abell AN, Ancrile BB, Johnson GL, Marchuk DA (2005) CCM1 and CCM2 protein interactions in cell signaling: implications for cerebral cavernous malformations pathogenesis. Hum Mol Genet 14:2521–2531
Acknowledgements
The authors thank the patients and their family for their cooperation and MRC Holland for developing the MLPA kits. Ute Felbor receives an Emmy Noether-grant from the Deutsche Forschungsgemeinschaft (Fe 432/6-5) and Sonja Stahl a stipend from the Graduiertenkolleg 1048.
Author information
Authors and Affiliations
Corresponding author
Additional information
Comments
Comment for Publication NSR-08-06-0117.R1
Hildegard Kehrer-Sawatzki, Ulm, Germany
The results reported by Gaetzner et al. are important in the context of mutation screening in families or sporadic cases suffering from recurrent headaches, seizures, and hemorrhagic stroke attributable to cerebral cavernous malformations. These lesions are frequently caused by mutations in one of the three CCM genes, CCM1, CCM2, or CCM3. Gaetzner et al. successfully applied the MLPA technique and identified a deletion of the CCM1 gene in a family with several affected members. Since smaller and larger deletions are often difficult or even impossible to identify by the analysis of polymorphic markers, the MLPA technique applied in this study proved to be an efficient method to identify such alterations unambiguously. Thus, the MLPA technique is an important addition to the current mutation detection protocols if sequencing of exons failed to reveal mutations. The manuscript is written very well and the results are presented in a clear and illustrative manner.
Comments
Comm f Publication NSR-08-06-0117.R1
Hidetoshi Kasuya, Tokyo, Japan
The exact mechanism of pathogenesis for familial cavernous malformation is not known. Mutations at three loci (CCM1, CCM2, and CCM3) have been shown in familial cavernous malformation. Three CCM genes likely act through the same molecular pathway because familial cavernous malformations caused by different gene mutations are pathologically and phenotypically indistinguishable. There is growing evidence that CCM1 may play a role in regulating ß1 integrin-mediated angiogenesis through this product, which is involved with a bidirectional signaling pathway between the extracellular matrix and the cytoskeleton that uses an integrin-mediated cascade [1]. Many mutations have been reported in CCM1: frameshifts, nonsense mutations, changes in the invariant splice junctions, missense mutations, and 84-base pair deletion [2]. MLPA allows the detection of midsize alterations by simultaneously screening for the loss or duplication of up to 50 target sequences. By using this newly developed technique, Gaetzner et al. successfully present the first report on a CCM1 gene deletion larger than the entire coding region, which would escape detection by the techniques reported before. 1. Dashti SR, Hoffer A, Hu YC, Selman WR (2006) Molecular genetics of familial cerebral cavernous malformations. Neurosurg Focus 21(1):E2 2. Verlaan DJ, Davenport WJ, Stefan H, Sure U, Siegel AM, Rouleau GA (2002) Cerebral cavernous malformations: Mutations in Krit1. Neurology 58: 853-8574
Comments
Comment to NSR-08-06-0117.R1
CCM1 gene deletion identified by MLPA in cerebral cavernous malformation
Dietmar Krex, Dresden, Germany
The pathogenesis of familial cerebral cavernous malformation is based on genetic variants within three genes, CCM1, -2, and -3, which is in contrast to most other cerebral vascular malformations where disease-causing genes still have to be determined. Therefore, predictive testing of at-risk relatives is possible by the analysis of blood samples; a goal that has to be achieved, for instance, for arterio-venous malformations or intracranial aneurysms.
However, valid diagnostics are hampered, as genetic variants within CCM genes not only comprise various mutations but also large genomic deletions, which might lead to false negative results by standard sequencing techniques. Gaetzner et al. show that, by using the multiplex ligation-dependent probe amplification gene dosage assay (MLPA), this particular shortcoming of missing genomic deletions can be overcome, making the analysis more accurate. As MLPA is an established technique based on commercially available kits, it can be widely and easily used for improving the predictive value of genetic testing. We are looking forward to results from larger cohorts tested in that comprehensive manner.
Rights and permissions
About this article
Cite this article
Gaetzner, S., Stahl, S., Sürücü, O. et al. CCM1 gene deletion identified by MLPA in cerebral cavernous malformation. Neurosurg Rev 30, 155–160 (2007). https://doi.org/10.1007/s10143-006-0057-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10143-006-0057-1