Abstract
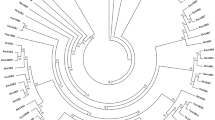
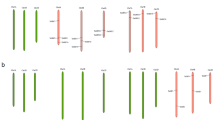
Maintenance of growth is important for sustaining yield under stress conditions. Hence, identification of genes involved in cell division and growth under abiotic stress is utmost important. Ras-related nuclear protein (Ran) is a small GTPase required for nucleocytoplasmic transport, mitotic progression, and nuclear envelope assembly in plants. In the present study, two Ran GTPase genes TaRAN1 and TaRAN2 were identified though genome-wide analysis in wheat (T. aestivum). Comparative analysis of Ran GTPases from wheat, barley, rice, maize, sorghum, and Arabidopsis revealed similar gene structure within phylogenetic clades and highly conserved protein structure. Expression analysis from expVIP platform showed ubiquitous expression of TaRAN genes across tissues and developmental stages. Under biotic and abiotic stresses, TaRAN1 expression was largely unaltered, while TaRAN2 showed stress specific response. In qRT-PCR analysis, TaRAN1 showed significantly higher expression as compared to TaRAN2 in shoot and root at seedling, vegetative, and reproductive stages. During progressive drought stress, TaRAN1 and TaRAN2 expression increase during early stress and restored to control level expression at higher stress levels in shoot. The steady-state level of transcripts was maintained to that of control in roots under drought stress. Under cold stress, expression of both the TaRAN genes decreased significantly at 3 h and became similar to control at 6 h in shoots, while salt stress significantly reduced the expression of TaRAN genes in shoots. The analysis suggests differential regulation of TaRAN genes under developmental stages and abiotic stresses. Delineating the molecular functions of Ran GTPases will help unravel the mechanism of stress induced growth inhibition in wheat.







Similar content being viewed by others
References
Barrs HD, Weatherley PE (1962) A re-examination of the relative turgidity technique for estimating water deficits in leaves. Aust J Biol Sci 15(3):413–428. https://doi.org/10.1071/BI9620413
Borrill P, Ramirez-Gonzalez R, Uauy C (2016) expVIP: a customizable RNA-seq data analysis and visualization platform. Plant Physiol 170:2172–2186. https://doi.org/10.1104/pp.15.01667
Chen N, Xu Y, Wang X, Du C, Du J, Yuan M, Xu Z, Chong K (2011) OsRAN2, essential for mitosis, enhances cold tolerance in rice by promoting export of intranuclear tubulin and maintaining cell division under cold stress. Plant Cell Environ 34:52–64. https://doi.org/10.1111/j.1365-3040.2010.02225.x
Clarke PR, Zhang C (2008) Spatial and temporal coordination of mitosis by Ran GTPase. Nat Rev Mol Cell Biol 9:464–477. https://doi.org/10.1038/nrm2410
Finn RD, Clements J, Arndt W, Miller BL, Wheeler TJ, Schreiber F, Bateman A, Eddy SR (2015) HMMER web server: 2015 update. Nucleic Acids Res 43:W30–W38. https://doi.org/10.1093/nar/gkv397
Haizel T, Merkle T, Pay A, Fejes E, Nagy F (1997) Characterization of proteins that interact with the GTP-bound form of the regulatory GTPase Ran in Arabidopsis. Plant J 11:93–103. https://doi.org/10.1046/j.1365-313x.1997.11010093.x
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31:1296–1297. https://doi.org/10.1093/bioinformatics/btu817
Jiang SY, Ramachandran S (2006) Comparative and evolutionary analysis of genes encoding small GTPases and their activating proteins in eukaryotic genomes. Physiol Genomics 24:235–251. https://doi.org/10.1152/physiolgenomics.00210.2005
Kahn RA, Der CJ, Bokoch GM (1992) The ras superfamily of GTP-binding proteins: guidelines on nomenclature. FASEB J 6:2512–2513. https://doi.org/10.1096/fasebj.6.8.1592203
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol 35:1547–1549
Letunic I, Doerks T, Bork P (2015) SMART: recent updates, new developments and status in 2015. Nucleic Acids Res 43:D257–D260. https://doi.org/10.1093/nar/gku949
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT Method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Lounsbury KM, Richards SA, Carey KL, Macara IG (1996) Mutations within the Ran/TC4 GTPase. Effects on regulatory factor interactions and subcellular localization. J Biol Chem 271:32834–32841. https://doi.org/10.1074/jbc.271.51.32834
Lycett G (2008) The role of Rab GTPases in cell wall metabolism. J Exp Bot 59:4061–4074. https://doi.org/10.1093/jxb/ern255
Ma L, Hong Z, Zhang Z (2007) Perinuclear and nuclear envelope localizations of Arabidopsis Ran proteins. Plant Cell Rep 26:1373–1382. https://doi.org/10.1007/s00299-007-0367-y
Molendijk AJ, Ruperti B, Palme K (2004) Small GTPases in vesicle trafficking. Curr Opin Plant Biol 7:694–700. https://doi.org/10.1016/j.pbi.2004.09.014
Moore MS (1998) Ran and nuclear transport. J Biol Chem 273:22857–22860. https://doi.org/10.1074/jbc.273.36.22857
Nagawa S, Xu T, Lin D, Dhonukshe P, Zhang X, Friml J, Scheres B, Fu Y, Yang Z (2012) ROP GTPase-dependent actin microfilaments promote PIN1 polarization by localized inhibition of clathrin-dependent endocytosis. PLoS Biol 10:e1001299. https://doi.org/10.1371/journal.pbio.1001299
Nibau C, Wu HM, Cheung AY (2006) RAC/ROP GTPases: 'hubs' for signal integration and diversification in plants. Trends Plant Sci 11:309–315. https://doi.org/10.1016/j.tplants.2006.04.003
Paolacci AR, Tanzarella OA, Porceddu E, Ciaffi M (2009) Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol Biol 10:11. https://doi.org/10.1186/1471-2199-10-11
Quimby BB, Dasso M (2003) The small GTPase Ran: interpreting the signs. Curr Opin Cell Biol 15:338–344. https://doi.org/10.1016/s0955-0674(03)00046-2
Ren M, Drivas G, D'Eustachio P, Rush MG (1993) Ran/TC4: a small nuclear GTP-binding protein that regulates DNA synthesis. J Cell Biol 120:313–323. https://doi.org/10.1083/jcb.120.2.313
Rojas AM, Fuentes G, Rausell A, Valencia A (2012) The Ras protein superfamily: evolutionary tree and role of conserved amino acids. J Cell Biol 196:189–201. https://doi.org/10.1083/jcb.201103008
Takai Y, Sasaki T, Matozaki T (2001) Small GTP-binding proteins. Physiol Rev 81:153–208. https://doi.org/10.1152/physrev.2001.81.1.153
Toma-Fukai S, Shimizu T (2019) Structural insights into the regulation mechanism of small GTPases by GEFs. Molecules 24(18):3308. https://doi.org/10.3390/molecules24183308
Vernoud V, Horton AC, Yang Z, Nielsen E (2003) Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol 131:1191–1208. https://doi.org/10.1104/pp.013052
Wang L, Ruan YL (2013) Regulation of cell division and expansion by sugar and auxin signaling. Front Plant Sci 4:163. https://doi.org/10.3389/fpls.2013.00163
Wang X, Xu Y, Han Y, Bao S, Du J, Yuan M, Xu Z, Chong K (2006) Overexpression of RAN1 in rice and Arabidopsis alters primordial meristem, mitotic progress, and sensitivity to auxin. Plant Physiol 140:91–101. https://doi.org/10.1104/pp.105.071670
Wu JY, Shen JR, Mao XZ, Liu KD, Wei LP, Liu PW, Yang GS (2007) Isolation and analysis of differentially expressed genes in dominant genic male sterility (DGMS) Brassica napus L. using substractive PCR and cDNA microarray. Plant Sci 172:204–211
Xu P, Cai W (2014) RAN1 is involved in plant cold resistance and development in rice (Oryza sativa). J Exp Bot 65:3277–3287. https://doi.org/10.1093/jxb/eru178
Xu P, Zang A, Chen H, Cai W (2016) The small G protein AtRAN1 regulates vegetative growth and stress tolerance in Arabidopsis thaliana. PLoS One 11:e0154787. https://doi.org/10.1371/journal.pone.0154787
Yang Z (2002) Small GTPases: versatile signaling switches in plants. Plant Cell 14(Suppl):S375–S388. https://doi.org/10.1105/tpc.001065
Yu CS, Chen YC, Lu CH, Hwang JK (2006) Prediction of protein subcellular localization. Proteins 64:643–651. https://doi.org/10.1002/prot.21018
Zang AP, Xu XJ, Neill S, Cai WM (2010) Overexpression of OsRAN2 in rice and Arabidopsis renders trans-genic plants hypersensitive to salinity and osmotic stress. J Exp Bot 61:777–789. https://doi.org/10.1093/jxb/erp341
Zhao Q, Leung S, Corbett AH, Meier I (2006) Identification and characterization of the Arabidopsis orthologs of nuclear transport factor 2, the nuclear import factor of Ran. Plant Physiol 140:869–878. https://doi.org/10.1104/pp.105.075499
The International Wheat Genome Sequencing Consortium (IWGSC), Appels R, Eversole K, Stein N, Feuillet C, Keller B, Rogers J, Pozniak CJ, Choulet F, Distelfeld A, Poland J, Ronen G. et al (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361(6403):eaar7191. https://doi.org/10.1126/science.aar7191
Acknowledgments
SC gratefully acknowledges ICAR for JRF fellowship.
Funding
This study was funded by ICAR-National Institute for Plant Biotechnology, New Delhi.
Author information
Authors and Affiliations
Contributions
Experimentation by SC; data analysis and manuscript editing by M; in silico analysis by SKM and MD; conceptualization by MD; manuscript writing JCP and MD. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Choudhury, S., Mansi, Muthusamy, S.K. et al. Genome-wide identification of Ran GTPase family genes from wheat (T. aestivum) and their expression profile during developmental stages and abiotic stress conditions. Funct Integr Genomics 21, 239–250 (2021). https://doi.org/10.1007/s10142-021-00773-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-021-00773-0