Abstract
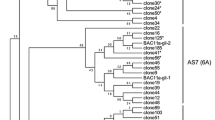
The complete set of unique γ-gliadin genes is described for the wheat cultivar Chinese Spring using a combination of expressed sequence tag (EST) and Roche 454 DNA sequences. Assemblies of Chinese Spring ESTs yielded 11 different γ-gliadin gene sequences. Two of the sequences encode identical polypeptides and are assumed to be the result of a recent gene duplication. One gene has a 3′ coding mutation that changes the reading frame in the final eight codons. A second assembly of Chinese Spring γ-gliadin sequences was generated using Roche 454 total genomic DNA sequences. The 454 assembly confirmed the same 11 active genes as the EST assembly plus two pseudogenes not represented by ESTs. These 13 γ-gliadin sequences represent the complete unique set of γ-gliadin genes for cv Chinese Spring, although not ruled out are additional genes that are exact duplications of these 13 genes. A comparison with the ESTs of two other hexaploid cultivars (Butte 86 and Recital) finds that the most active genes are present in all three cultivars, with exceptions likely due to too few ESTs for detection in Butte 86 and Recital. A comparison of the numbers of ESTs per gene indicates differential levels of expression within the γ-gliadin gene family. Genome assignments were made for 6 of the 13 Chinese Spring γ-gliadin genes, i.e., one assignment from a match to two γ-gliadin genes found within a tetraploid wheat A genome BAC and four genes that match four distinct γ-gliadin sequences assembled from Roche 454 sequences from Aegilops tauschii, the hexaploid wheat D-genome ancestor.





Similar content being viewed by others
References
Altenbach SB, Vensel WH, Dupont FM (2010) Analysis of expressed sequence tags from a single wheat cultivar facilitates interpretation of tandem mass spectrometry data and discrimination of gamma gliadin proteins that may play different functional roles in flour. BMC Plant Biol 10:7
Anderson OD (2009) EST mining for structure and expression of genes in the region of the wheat high-molecular-weight glutenin loci. Genome 52:726–740
Anderson OD, Greene FC (1997) The alpha-gliadin gene family. II. DNA and protein sequence variation, subfamily structure, and origins of pseudogenes. Theor Appl Genet 95:59–65
Anderson OD, Hsia CC, Torres V (2001) The wheat gamma-gliadin genes: characterization of ten new sequences and further understanding of gamma-gliadin gene family structure. Theor Appl Genet 103:323–330
Anderson OD, Dong L, Huo N, Gu YQ (2012) A new class of wheat gliadin genes and proteins. PLoS One 7:e52139
Armstrong MJ, Hegade VS, Robins G (2012) Advances in coeliac disease. Curr Opin Gastroenterol 28:104–112
Chao S, Lazo GR, You F, Crossman CC, Hummel DD, Lui N, Laudencia-Chingcuanco D, Anderson JA, Close TJ, Dubcovsky J, Gill BS, Gill KS, Gustafson JP, Kianian SF, Lapitan NLV, Nguyen HT, Sorrells ME, McGuire PE, Qualset CO, Anderson OD (2006) Use of a large-scale Triticeae expressed sequence tag resource to reveal gene expression profiles in hexaploid wheat (Triticum aestivum L.). Genome 49:531–544
Ferrante P, Masci S, D’Ovidio R, Lafiandra D, Volpi C, Mattei B (2006) A proteomic approach to verify in vivo expression of a novel γ-gliadin containing an extra cysteine residue. Proteomics 6:1908–1914
Gao S, Gu Y, Wu J, Coleman-Derr D, Huo N, Crossman C, Jia J, Zuo Q, Ren Z, Anderson OD, Kong X (2007) Rapid evolution and complex structural organization in genomic regions harboring multiple prolamin genes in the polyploid wheat genome. Plant Mol Biol 65:189–203
Krzyzosiak WJ, Sobczak K, Wojciechowska M, et al. (2012) Triplet repeat RNA structure and its role as pathogenic agent and therapeutic target. Nucleic Acids Res 40:11–26
Lafiandra D, Colaprico G, Kasarda DD, Porceddu E (1987) Null alleles for gliadin blocks in bread and durum wheat cultivars. Theor Appl Genet 74:610–616
Lazo GR, Chao S, Hummel DD, Edwards H, Crossman CC, Lui N, Matthews DE, Carollo VL, Hane DL, You FM, Butler GE, Miller RE, Close TJ, Peng JH, Laptian NLV, Gustafson JP, Qi LL, Echalier B, Gill BS, Dilbirlgi M, Randhawa HS, Gill KS, Greene RA, Sorrells ME, Akhunov ED, Dvorak J, Linkiewicz AM, Dubcovsky J, Hossain KG, Kalavacharla V, Kianian SF, Mahmoud AA, Miftahudin MXF, Conley EJ, Anderson JA, Pathan MS, Nguyen HT, McGuire PE, Qualset CO, Anderson OD (2004) Development of an expressed sequence tag (EST) resource for wheat (Triticum aestivum L.): EST generation, unigene analysis, probe selection and bioinformatics for a 16,000-locus bin-delineated map. Genetics 168:585–593
MacRitchie F (1992) Physicochemical properties of wheat proteins in relation to functionality. Adv Food Nutr Res 36:1–87
Mitrovich QM, Anderson P (2005) mRNA surveillance of expressed pseudogenes in C. elegans. Curr Biol 15:963–967
Müller S, Wieser H (1997) The location of disulphide bonds in monomeric gamma-type gliadins. J Cereal Sci 26:169–176
Payne PI (1987) Genetics of wheat storage proteins and the effects of allelic variation on bread-making quality. Ann Rev Plant Physiol 38:141–153
Pistón F, Dorado G, Martín A, Barro F (2006) Cloning of nine γ-gliadin mRNAs (cDNAs) from wheat and the molecular characterization of comparative transcript levels of γ-gliadin subclasses. J Cereal Sci 43:120–128
Qi PF, Wei YM OT, Chen Q, Tan X, Zheng YL (2009) The gamma-gliadin multigene family in common wheat (Triticum aestivum) and its closely related species. BMC Genomics 10:168
Rafalski JA (1986) Structure of wheat gamma-gliadin genes. Gene 43:221–229
Sabelli PA, Shewry PR (1991) Characterization and organization of gene families at the Gli-1 loci of bread and durum wheats by restriction fragment analysis. Theor Appl Genet 83:209–227
Shewry PR, Halford NG, Lafiandra D (2003) Genetics of wheat gluten proteins. Adv Genet 49:111–184
Wang S, Shen X, Ge P, Li J, Subburaj S, Li X, Zeller FJ, Hsam SLK, Yan Y (2012) Molecular characterization and dynamic expression patterns of two types of γ-gliadin genes from Aegilops and Triticum species. Theor Appl Genet 125:1371–1384
Acknowledgments
The research was supported by USDA-ARS project 5325-21000-015 and National Science Foundation grant 0701916.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. S1
γ-Gliadin gene sequence source. The sequence structure of the 11 Chinese Spring active γ-gliadin genes is diagrammed. Start (ATG) and stop (TGA) codon positions are marked. Genes CS-γ6a and CS-γ6b are diagrammed together since the flanking DNA is difficult to distinguish between the two nearly identical genes. Gray, non-coding flanking DNA; dark blue, coding sequence with at least two 454 reads covering those sections; light blue, only one 454 read; yellow, no 454 reads (JPEG 16 kb)
High Resolution Image
(TIFF 26 kb)
Supplementary Fig. S2
Origin of an anomalous carboxy terminus of a γ-gliadin. The CS-γ5 polypeptide has a different carboxy terminus from other known γ-gliadins. Shown is how a complex duplication at the 3′ end of the coding sequence changed the reading frame. The 3′ end of the CS-γ5 reading frame is compared with CS-γ2. Stop codons for both polypeptides are boxed. Encoding amino acids are indicated above and below the two DNA sequences. Brackets above and below the CS-γ5 sequence indicate duplications (JPEG 52 kb)
High Resolution Image
(TIFF 681 kb)
Supplementary File S1
A fasta file of the extended γ-gliadin sequences for hexaploid wheat cultivar Chinese Spring. Sequences are a consensus of EST and 454 sequences, as described in “Materials and methods.” Sequences gamma-13 and gamma-14 are pseudogenes whose sequences are derived completely from 454 reads (FAS 28 kb)
Supplementary File S2
A fasta file of the γ-gamma consensus sequences from ESTs of hexaploid wheat cultivars Butte 86 and Recital (FAS 17 kb)
Rights and permissions
About this article
Cite this article
Anderson, O.D., Huo, N. & Gu, Y.Q. The gene space in wheat: the complete γ-gliadin gene family from the wheat cultivar Chinese Spring. Funct Integr Genomics 13, 261–273 (2013). https://doi.org/10.1007/s10142-013-0321-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-013-0321-8