Abstract
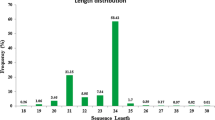
The small RNA transcriptomes of bread wheat and its emerging model Brachypodium distachyon were obtained by using deep sequencing technology. Small RNA compositions were analyzed in these two species. In addition to 70 conserved microRNAs (miRNAs) from 25 families, 23 novel wheat miRNAs were identified. For Brachypodium, 12 putative miRNAs were predicted from a limited number of expressed sequence tags, of which one was a potential novel miRNA. Also, 94 conserved miRNAs from 28 families were identified in this species. Expression validation was performed for several novel wheat miRNAs. RNA ligase-mediated 5′ rapid amplification of complementary DNA ends experiments demonstrated their capability to cleave predicted target genes including three disease-resistant gene analogs. Differential expression of miRNAs was observed between Brachypodium vegetative and reproductive tissues, suggesting their different roles at the two growth stages. Our work significantly increases the novel miRNA numbers in wheat and provides the first set of small RNAs in B. distachyon.




Similar content being viewed by others
References
Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121:207–221
Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths-Jones S, Marshall M, Matzke M, Ruvkun G, Tuschl T (2003) A uniform system for microRNA annotation. RNA 9:277–279
Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a MicroRNA and its APETALA2-like target genes. Plant Cell 15:2730–2741
Barakat A, Wall PK, Diloreto S, Depamphilis CW, Carlson JE (2007) Conservation and divergence of microRNAs in Populus. BMC Genomics 8:481
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Bartel B, Bartel DP (2003) MicroRNAs: at the root of plant development? Plant Physiol 132:709–717
Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK (2005) Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123:1279–1291
Boualem A, Laporte P, Jovanovic M, Laffont C, Plet J, Combier JP, Niebel A, Crespi M, Frugier F (2008) MicroRNA166 controls root and nodule development in Medicago truncatula. Plant J 54:876–887
Bourbonnais R, Paice MG (1990) Oxidation of non-phenolic substrates. An expanded role for laccase in lignin biodegradation. FEBS Lett 267:99–102
Bouton JH (2007) Molecular breeding of switchgrass for use as a biofuel crop. Curr Opin Genet Dev 17:553–558
Carrington JC, Ambros V (2003) Role of microRNAs in plant and animal development. Science 301:336–338
Chapman EJ, Carrington JC (2007) Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet 8:884–896
Chen X (2004) A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303:2022–2025
Chen ZJ (2007) Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Annu Rev Plant Biol 58:377–406
Draper J, Mur LAJ, Jenkins G, Ghosh-Biswas GC, Bablak P, Hasterok R, Routledge APM (2001) Brachypodium distachyon. A new model system for functional genomics in grasses. Plant Physiol 127:1539–1555. doi:10.1104/pp.010196
Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, Carrington JC (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE 2:e219
Gordon D, Desmarais C, Green P (2001) Automated finishing with autofinish. Genome Res 11:614–625
Hamilton A, Voinnet O, Chappell L, Baulcombe D (2002) Two classes of short interfering RNA in RNA silencing. EMBO J 21:4671–4679
Herr AJ (2005) Pathways through the small RNA world of plants. FEBS Lett 579:5879–5888
Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14:787–799
Jurka J (2000) Repbase update: a database and an electronic journal of repetitive elements. Trends Genet 9:418–420
Kurihara Y, Watanabe Y (2004) Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci U S A 101:12753–12758
Lau NC, Lim LP, Weinstein EG, Bartel DP (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294:858–862
Lauter N, Kampani A, Carlson S, Goebel M, Moose SP (2005) microRNA172 down-regulates glossy15 to promote vegetative phase change in maize. Proc Natl Acad Sci 102:9412–9417. doi:10.1073/pnas.0503927102
Lindow M, Krogh A (2005) Computational evidence for hundreds of non-conserved plant microRNAs. BMC Genomics 6:119
Lippman Z, Martienssen R (2004) The role of RNA interference in heterochromatic silencing. Nature 431:364–370
Liu B, Li P, Li X, Liu C, Cao S, Chu C, Cao X (2005) Loss of function of OsDCL1 affects microRNA accumulation and causes developmental defects in rice. Plant Physiol 139:296–305. doi:10.1104/pp.105.063420
Llave C, Xie Z, Kasschau KD, Carrington JC (2002) Cleavage of scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297:2053–2056
Lu C, Jeong DH, Kulkarni K, Pillay M, Nobuta K, German R, Thatcher SR, Maher C, Zhang L, Ware D, Liu B, Cao X, Meyers BC, Green PJ (2008) Genome-wide analysis for discovery of rice microRNAs reveals natural antisense microRNAs (nat-miRNAs). Proc Natl Acad Sci 105:4951–4956. doi:10.1073/pnas.0708743105
Meyers BC, Axtell MJ, Bartel B, Bartel DP, Baulcombe D, Bowman JL, Cao X, Carrington JC, Chen X, Green PJ, Griffiths-Jones S, Jacobsen SE, Mallory AC, Martienssen RA, Poethig RS, Qi Y, Vaucheret H, Voinnet O, Watanabe Y, Weigel D, Zhu JK (2008) Criteria for annotation of plant microRNAs. Plant Cell 20:3186-3190
Millar AA, Waterhouse PM (2005) Plant and animal microRNAs: similarities and differences. Funct Integr Genomics 5:129–135
Mlotshwa S, Yang Z, Kim Y, Chen X (2006) Floral patterning defects induced by Arabidopsis APETALA2 and microRNA172 expression in Nicotiana benthamiana. Plant Mol Biol 61:781–793
Oh MH, Wang X, Kota U, Goshe MB, Clouse SD, Huber SC (2009) Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis. Proc Natl Acad Sci U S A 106:658–663
Opanowicz M, Vain P, Draper J, Parker D, Doonan JH (2008) Brachypodium distachyon: making hay with a wild grass. Trends Plant Sci 13:172–177
Ori N, Cohen AR, Etzioni A, Brand A, Yanai O, Shleizer S, Menda N, Amsellem Z, Efroni I, Pekker I, Alvarez JP, Blum E, Zamir D, Eshed Y (2007) Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat Genet 39:787–791
Qi Y, He X, Wang XJ, Kohany O, Jurka J, Hannon G (2006) Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature 443:1008–1012
Rajagopalan R, Vaucheret H, Trejo J, Bartel DP (2006) A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev 20:3407–3425. doi:10.1101/gad.1476406
Subramanian S, Fu Y, Sunkar R, Barbazuk WB, Zhu JK, Yu O (2008) Novel and nodulation-regulated microRNAs in soybean roots. BMC Genomics 9:160
Sunkar R, Girke T, Jain PK, Zhu JK (2005) Cloning and characterization of microRNAs from rice. Plant Cell 17:1397–1411
Sunkar R, Zhou X, Zheng Y, Zhang W, Zhu JK (2008) Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol 8:25
Vaucheret H (2006) Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev 20:759–771. doi:10.1101/gad.1410506
Vazquez F (2006) Arabidopsis endogenous small RNAs: highways and byways. Trends Plant Sci 11:460–468
Vogel JP, Gu YQ, Twigg P, Lazo GR, Laudencia-Chingcuanco D, Hayden DM, Donze TJ, Vivian LA, Stamova B, Coleman-Derr D (2006) EST sequencing and phylogenetic analysis of the model grass Brachypodium distachyon. Theor Appl Genet 113:186–195
Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2:E104
Xie K, Wu C, Xiong L (2006) Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol 142:280–293. doi:10.1104/pp.106.084475
Yao Y, Guo G, Ni Z, Sunkar R, Du J, Zhu JK, Sun Q (2007) Cloning and characterization of microRNAs from wheat (Triticum aestivum L.). Genome Biol 8:R96
Zhao T, Li G, Mi S, Li S, Hannon GJ, Wang XJ, Qi Y (2007) A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev 21:1190–1203. doi:10.1101/gad.1543507
Zilberman D, Cao X, Johansen LK, Xie Z, Carrington JC, Jacobsen SE (2004) Role of Arabidopsis ARGONAUTE4 in RNA-directed DNA methylation triggered by inverted repeats. Curr Biol 14:1214–1220
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415
Acknowledgements
We thank Dr. Charles Leseberg for critical reading of this manuscript. We are grateful to Professor Rudi Appels and Dr. Wujun Ma who provided constructive suggestions during the revision of the manuscript. This work was supported in part by the National HITECH Research and Development Program of China (“863” program, #2006AA10A104), Special Research Grant for Central Governmental Nonprofit Research Institutes (Yuan-Suo-Zhang-Ji-Jin), and National Basic Research Program (“973” program, #2004CB117200) of China.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Wei, Cai, and Zhang contribute equally to this work.
Rights and permissions
About this article
Cite this article
Wei, B., Cai, T., Zhang, R. et al. Novel microRNAs uncovered by deep sequencing of small RNA transcriptomes in bread wheat (Triticum aestivum L.) and Brachypodium distachyon (L.) Beauv. Funct Integr Genomics 9, 499–511 (2009). https://doi.org/10.1007/s10142-009-0128-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10142-009-0128-9