Abstract
Background
Signet ring cell carcinoma (SRCC) is a particular histologic variant of gastric cancer (GC). However, the critical factor related to the aggressive characteristics of SRCC has not been determined.
Methods
We collected surgically resected tissues from 360 GC patients in the Kumamoto University cohort and generated survival curves via the Kaplan–Meier method. In vitro, we identified the specific transcript variant of MUC20 in SRCC cells by direct sequencing and investigated the role of MUC20 in GC progression using GC cells with MUC20 silencing and forced expression. In vivo, we examined chemoresistance using MUC20 variant 2 (MUC20v2)-overexpressing non-SRCC cells to construct a xenograft mouse model.
Results
We analyzed a comprehensive GC cell line database to identify the specifically expressed genes in gastric SRCC. We focused on MUC20 and investigated its role in GC progression. Survival analysis revealed that GC patients with high MUC20 expression exhibited a poor prognosis and that MUC20 expression was significantly correlated with SRCC histological type. Moreover, we found that gastric SRCC cells specifically expressed MUC20v2, which was dominantly expressed in the cytoplasm. Silencing MUC20v2 caused cell death with characteristic morphological changes in gastric SRCC cells. To further determine the types of cell death, we examined apoptosis, pyroptosis and ferroptosis by detecting cleaved PARP, gasdermin E-N-terminal (GSDME-N), and lipid reactive oxygen species (ROS) levels, respectively. We found that apoptosis and pyroptosis occurred in MUC20-silenced gastric SRCC cells. In addition, MUC20v2-overexpressing GC cells exhibited chemoresistance to cisplatin (CDDP) and paclitaxel (PTX). RNA sequencing revealed that the pathways involved in intracellular calcium regulation were significantly upregulated in MUC20v2-overexpressing GC cells. Notably, forced expression of MUC20v2 in the cytoplasm of GC cells led to the maintenance of mitochondrial calcium homeostasis and mitochondrial membrane potential (MMP), which promoted cell survival and chemoresistance by suppressing apoptosis and pyroptosis. Finally, we investigated the significance of MUC20v2 in a xenograft model treated with CDDP and showed that MUC20v2 overexpression caused chemoresistance by inhibiting cell death.
Conclusion
These findings highlight the novel functions of MUC20v2, which may confer cell survival and drug resistance in GC cells.
Significance
MUC20v2 protects GC cells from apoptosis and pyroptosis by maintaining mitochondrial calcium levels and mitochondrial membrane potential and subsequently induces drug resistance.
Similar content being viewed by others
Introduction
Gastric cancer (GC) is the most common malignancy in Eastern Asia and the fifth most frequently diagnosed cancer worldwide, accounting for over 1,000,000 new cancer cases in 2018. Due to its high mortality rate, GC remains the third leading cause of cancer-related death worldwide [1]. Histologically, GC is classified into intestinal and diffuse types according to Lauren’s classification, and the diffuse type shows a worse prognosis than the intestinal type [2, 3]. However, with the further classification of GC types by microscopic characteristics, the World Health Organization (WHO) classification divides GC into well-differentiated, moderately differentiated, poorly differentiated adenocarcinoma, and signet ring cell carcinoma (SRCC) [4]. Notably, the WHO classification defines SRCC as GC containing at least 50% of SRCC cells in pathologic specimens [5, 6]. Although declines in GC incidence have been reported, the proportion of GC cases classified as gastric SRCC has increased worldwide [7]. In addition, patients with advanced gastric SRCC show a worse prognosis and lower chemosensitivity than those with other GC types [8,9,10].
In terms of histological imaging, SRCC cells are uniquely characterized as rounded cells with intracellular abundant mucin (MUC), misplaced nuclei, and crescent-shaped hyperchromatic nuclei. Additionally, the cohesion of SRCC cells is worse than that of the other types. In particular, MUC production is a specific biological hallmark of SRCC. Although MUC2 and MUC5AC have been used for the differential diagnosis of histological types in various cancers [11,12,13], mucin staining is insufficient to distinguish between gastric SRCC and non-SRCC [14]. In addition, comprehensive genomic analysis revealed that CLDN18-ARHGAP26/6 fusion has been frequently observed in SRCC patients, and this fusion is correlated with chemotherapy resistance and a poor prognosis [15]. However, the highly expressed molecules and intracellular signaling related to the aggressive characteristics of SRCC have not been determined. Therefore, our study aimed to identify the critical molecule regulating the unique characteristics of SRCC cells.
Materials and methods
Cell lines and cell culture
Human GC cell lines were purchased from the RIKEN BioResource Center Cell Bank (MKN1, MKN7, KATO III, MKN45) and the Japanese Collection of Research Bioresources Cell Bank (NUGC3, NUGC4). All the cell lines tested negative for Mycoplasma using the e-Myco Mycoplasma PCR Detection Kit (#25,235, Cosmo bio, Japan) during the period of this study. These cell lines were cultured in RPMI 1640 medium containing 10% fetal bovine serum (FBS) and maintained at 37 °C in a humidified atmosphere containing 5% CO2.
TIMER 2.0
TIMER 2.0 (http://timer.cistrome.org/) is an online tool for analyzing gene expression between tumor tissue and normal tissue in the TCGA portal [16].
Kaplan–Meier plotter
Kaplan–Meier plotter (http://www.kmplot.com/analysis/) was used to analyze the impact of 54,675 genes on survival in 21 cancer types. Sources of the system database include the Gene Expression Omnibus (GEO), The European Genome-Phenome Archive (EGA), and the TCGA. This tool can be applied to perform survival curve analysis [17]. The database includes KLK7 expression data for 875 GC patients and MUC20 expression data for 631 GC patients. We used the median expression levels as the cut-off values for these patients.
Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR)
RNA was extracted from cultured cells using the RNeasy Mini Kit (#74,106, Qiagen, Hilden, Germany) according to the protocol recommended by the manufacturer. Complementary DNA (cDNA) was reverse transcribed from total RNA using SuperScript III, RNaseOUT, Recombinant Ribonuclease Inhibitor, Random Primers and Oligo(dT)12–18 Primer (Thermo Fisher Scientific, Tokyo, Japan). Quantitative reverse transcriptase (qRT)-PCR was performed using the TaqMan Probe method. mRNA expression was measured by qRT-PCR using the TaqMan probe (Roche Diagnostics, Basel, Switzerland), and the value for each gene was normalized to that for β-actin. mRNA expression was measured by qRT-PCR using the TaqMan probe (Roche Diagnostics, Basel, Switzerland), and the value for each gene was normalized to that for β-actin. All qRT-PCR experiments were performed using a LightCycler 480 System II (Roche Diagnostics). All qRT-PCR data are shown as the mean ± standard error of the mean. The MUC20 forward primer sequence was 5'-AGTGGGCAAAACAACTTCCTT-3′; the MUC20 reverse primer sequence was 5′-GCTTCCGAGGGGCTGTAG-3′ (Universal Probe Library Probes: 38#). The β-actin forward primer sequence was 5′-ATTGGCAATGAGCGGTT-3′, and the β-actin reverse primer sequence was 5′-CGTGGATGCCACAGGACT-3′ (Universal Probe Library Probes: #11).
siRNA transfection
MUC20 expression was downregulated by predesigned Silencer Select siRNAs directed against MUC20 (#1, s47302; #2, s47304; Cat# 4,427,037, Thermo Fisher Scientific), and a nontargeting siRNA (#4,390,843, Thermo Fisher Scientific) was used as a negative control. SRCC cells were transfected with annealed siRNAs using Lipofectamine RNAiMAX (#13,778–150, Thermo Fisher Scientific).
IF cytochemical analysis for MUC20
GC cells were cultured in 35 mm glass-bottomed dishes (Matsunami Glass Industry, Japan) for 24 h. After treatment with normal medium, MUC20 siRNA and antitumor drug were added for 24 h at 37 °C. For MUC20 staining, GC cells were stained with the membrane dye wheat germ agglutinin for 30 min in the dark at 37 °C, fixed with acetone for 10 min and blocked in 5% low-fat dry milk for 30 min. The cells were stained with primary antibodies against MUC20 overnight at 4 °C. For calcium staining, the cells were stained with Fluo-4 AM first for 15 min in dark at 37 °C. After 3 washes with PBS, the cells were stained with Rhod-2 AM for 15 min in the dark at 37 °C. Alexa Fluor 488-conjugated antibody was used as the secondary antibody, and the nuclei were counterstained with DAPI. The antibodies used are listed in (Supplementary Table 3). The stained cells were observed with an FV1200 laser scanning confocal microscope (Olympus, Japan). The cells with MUC20-positive expression (at least one-third of the cellular membrane) were quantified by counting the stained nuclei in five fields of view. The cells with calcium expression were also quantified by counting the cells in five fields of view.
Cytotoxicity assay
For cytotoxicity assays, cells were co-cultured with Incucyte® Cytotox Green Dye (Supplementary Table 3) for 72–96 h according to the manufacturer’s instructions. Images of stained cells were taken with an Incucyte S3 Live-Cell Analysis System (Essen BioScience, Germany). The green cells were quantified by Incucyte software, all groups included triplicate measurements, and the data are presented as the mean ± standard error of the mean.
Protein extraction from whole-cell lysates and western blotting
Protein extraction from whole-cell lysates and Western blotting were performed as previously described with minor modifications [18]. The cultured cells were washed twice with cold PBS and lysed in RIPA buffer containing 1 × Halt Protease and Phosphatase Inhibitor Cocktail (#78,442, Thermo Fisher Scientific). The lysate was sonicated for 1 s at 1 s intervals for a total period of 1 min and centrifuged at 15,000×g and 4 °C for 15 min. The supernatant was then collected as the whole-cell lysate. The protein samples were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes. For Western blotting, the membranes were blocked with 5% low-fat dry milk and incubated overnight at 4 °C with primary antibodies against MUC20, β-actin, caspase-3, cleaved caspase-3, cleaved PARP, and GSDME-N diluted in TBST. The signals were detected by incubation with rabbit secondary antibodies at room temperature for 1 h using an enhanced chemiluminescence (ECL) detection system (GE Healthcare, Chicago, Illinois, US). The antibodies used are listed in (Supplementary Table 3).
Flow cytometric analysis
The cell concentration was adjusted to 1 × 106 cells/mL in PBS containing 1% BSA. The cell suspensions were incubated with antibodies for 30 min at room temperature, washed with PBS containing 1% BSA and centrifuged twice, and the cell pellet was suspended in PBS. The antibodies used are listed in Supplementary Table 3. Flow cytometry was performed with FACSVerse and FACSAria III instruments (BD Biosciences, New Jersey, US). The flow cytometry data were analyzed using FlowJo 3.3 software (TreeStar).
Cell death assay
Supernatants containing floating apoptotic cells were collected, and the adherent cells were pelleted and combined with the floating cells. The collected cells were washed twice with PBS, and each pellet was resuspended in annexin V binding buffer at 1 × 107 cells/mL. The cells were stained with annexin V and 7-AAD and incubated in the dark at room temperature for 15 min. Annexin V binding buffer was added to each sample, and the cells were analyzed using a FACSVerse instrument (BD Biosciences). The antibodies used are listed in (Supplementary Table 3).
Lipid peroxidation assay
After cell seeding and treatment, the cells were harvested by trypsinization and preloaded with 5 µM C11-BODIPY 581/591 (Supplementary Table 3) in PBS for 1 h at 37 °C. The cells were assessed using a FACSVerse instrument with a 488 nm laser on a FITC detector.
Semiquantitative PCR and sequencing
RNA extraction and quantitative reverse transcription PCR followed the previous protocol that we described. The cDNA of GC cells was amplified by PCR using MUC20 primers (forward: 5'-ATTGGCTCACACCTCCACAG-3'; reverse: 5'-CTGTGACCAGAGCTCCACTG-3'). The PCR products were analyzed by electrophoresis on agarose gels. Detected bands were extracted by the QIAquick Gel Extraction Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s instructions. The purified DNA was sent for DNA sequencing on an ABI PRISM 3130 (Applied Biosystems™) using a BigDye Terminators ver. Cycle Sequencing Kit (Applied Biosystems™). The sequencing results were analyzed with GENETYX Ver. 13 software (Genetyx Corporation, Japan).
Construction of a stable cell line expressing MUC20
A plasmid containing the MUC20v2 precursor (Supplementary Table 3) was punched from ORIGENE (OriGene Technologies, Rockville, MD, USA). The open reading frame (ORF) of MUC20 was amplified by PCR using a forward primer containing the Sgf I site (5’-GAGGCGATCGCCATGGCCAAACCAGCACAGGGTGC-3’) and a reverse primer containing the MIuI site (5’-
GCGACGCGTGTCCTCACCACCACAGAGAGCCAG-3’) to append the cloning sites. The ORF was cloned into the pLenti-C-Myc-DDK-IRES-Puro Tagged Cloning Vector (Supplementary. Table 3) using AsiSI endonuclease (an isoschizomer of SgfI (#R0630S, New England Biolabs, Massachusetts, US) and MluI (#R3198S, New England Biolabs). Lentiviral particles were produced with Lentiviral High Titer Packaging Mix (Supplementary. Table 3) according to the manufacturer’s recommended protocol. Briefly, the destination vectors and lentiviral High Titer Packaging Mix were transfected into the Lenti-X™ 293 T cell line (#Z2180N, TaKaRa) using Lipofectamine 3000 (Supplementary. Table 3). The NUGC3 and MKN45 cell lines were cultured in complete medium containing viral particles with 4 µg/mL polybrene for 24 h. NUGC3 cells infected with lentivirus were selected by incubation with 2 µg/mL puromycin for 2 weeks. MKN45 cells infected with lentivirus were selected by incubation with 1.5 µg/mL puromycin for 2 weeks.
RNA-seq
RNA-seq was conducted as established in a previous study conducted at Kumamoto University [19]. RNA-seq was performed by the Liaison Laboratory Research Promotion Center (LILA) (Kumamoto University) as follows. The samples included two control cell lines (MKN45-MOCK and NUGC3-MOCK) and two overexpressed cell lines (MKN45-MUC20v2-OE and NUGC3-MUC20v2-OE). Total RNA was isolated using the RNeasy Mini Kit (#74,106, Qiagen), and an Agilent 2100 bioanalyzer (Agilent) was used to measure the total RNA concentration and purity. All the samples with an RNA integrity number (RIN) > 8.0 were used for sequencing. A NextSeq 500 platform (Illumina) was used for the analysis, and the data were converted to Fastq format. The quality of the data was determined by Trim galore (v0.5.0). The filtered reads were then mapped to the GRCh38 reference genome using STAR (v2.6.0). Transcripts per million mapped reads (TPM) values were calculated using RSEM. The differentially expressed genes (DEGs) between MUC20v2-overexpressing groups (NUGC3-MUC20v2-OE, MKN45-MUC20v2-OE) and control groups (NUGC3-MOCK, MKN45-MOCK) were screened using the Subio Platform software ver1.24 (Subio Inc., Amami, Japan). P value < 0.05 and |log2 FC|> 1.5) were set as cut-off standards and considered to indicate statistical significance. Functional and pathway enrichment analysis was performed via Metascape (https://metascape.org). The RNA-seq data were deposited in the DNA Data Bank of Japan (DDBJ) database under the accession number DRA012664.
Measurement of mitochondrial membrane depolarization and intracellular and mitochondrial calcium levels
The intracellular calcium and mitochondrial calcium levels of the indicated cells were determined according to the manufacturer’s instructions using a MitoProbe JC-1 Assay kit, Fluo-4 AM, Rhod-2, AM, respectively. In brief, the cells were incubated in the dark at room temperature for 30 min. The antibodies and dye used are listed in (Supplementary table 3). After staining, the fluorescence intensity of each sample was determined using a flow cytometer.
Animals
Six-week-old nude male BALB/c mice (Clea Japan, Shizuoka, Japan) were housed in a room with a stable temperature and humidity with a 12 h light/dark cycle. Both water and food were supplied ad libitum. All animal studies were conducted using protocols approved by the Institutional Animal Care and Use Committee of Kumamoto University (Approval Number: A2021-109).
In vivo chemoresistance assay
In vivo chemoresistance experiments were performed on six-week-old nude male BALB/c mice (Clea Japan, Shizuoka, Japan). The mice were subcutaneously transplanted with NUGC3-MOCK and NUGC3-MUC20v2-OE human cells (2.5 × 106). After 3 weeks, cisplatin (1.5 mg/kg) was injected intraperitoneally (i.p.) into the mice. The protocol used is described in Fig. 6A. The mice were euthanized on the second day after the last injection, and then the subcutaneous tumors were removed and weighed.
Human samples
Tissue samples were obtained from 340 patients with GC who treated at Kumamoto University Hospital between April 2005 and December 2016. All patients underwent curative gastrectomy, systemic chemotherapy, or conversion surgery and their resected samples were utilized. Clinical data including tumor stage and survival data were obtained from their medical records.
Immunohistochemical (IHC) staining and analysis
IHC staining was performed according to standard methods as described by our previous study [20]. The antibodies used are listed in (Supplementary Table 3). All samples were scored in a blinded manner by two independent investigators. MUC20 and KLK7 staining was scored according to the intensity and range of staining in cancer cells. The average positive staining intensity in cancer cells was determined, and an intensity score ranging from 1 (weak) to 2 (strong) was assigned. In addition, the average proportion of positively stained cancer cells was estimated, and a percentage score ranging from 1 to 4 was assigned: 1, < 25%; 2, 25–50%; 3, 50–75%; and 4, > 75% positive staining. The two scores were multiplied to define MUC20 and KLK7 expression as low (scores of 1–4) or high (scores of 5–8) (Fig. 1 C).
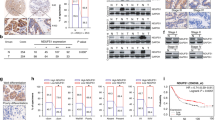
MUC20 is dominantly expressed in SRCC cells and is correlated with a poor prognosis in GC patients. A Genome-wide mRNA expression analysis of GC cell lines revealed significant DEGs in SRCC cells. B Overall survival curve analysis of GC patients stratified by KLK7 and MUC20 expression levels with the Kaplan–Meier plotter online database. P values indicating the significance of differences between the high and low expression groups were calculated using the log-rank test. C Representative IHC staining of KLK7 and MUC20 and quantification of KLK7-positive and MUC20-positive cells. D Overall survival curves of all patients with GC based on their KLK7 and MUC20 status. E Overall survival curves of patients with stage IV GC treated with chemotherapy based on their MUC20 status. F MUC20 expression levels in GC cell lines were assessed by qRT-PCR. G The expression of MUC20 in the NUGC3, MKN45 and NUGC4 cell lines was evaluated by Western blotting analysis. β-Actin was similarly analyzed as a loading control. H IF staining for markers of MUC20 and membrane dye wheat germ agglutinin together with nuclear staining with DAPI. I Cell death assay of GC cells treated with cisplatin for 24 h. Then, the cells were stained with annexin V and 7-AAD (#420,403, BioLegend). (n = 3 (F); scale bars, 200 um (C), 25 μm (D))
TUNEL staining
Paraffin-embedded sections of tissues obtained from the mice were deparaffinized and soaked in 0.85% NaCl. The In Situ BrdU-Red DNA Fragmentation (TUNEL) Assay Kit (Supplementary. Table 3) was used for TUNEL analysis. After treatment with the DNA-labeling solution, the samples were incubated overnight. The next day, the samples were treated with 100 µL of antibody solution for 30 min at room temperature, and the nuclei were counterstained with DAPI.
Study approval
This research was approved by the Medical Ethics Committee of Kumamoto University. Written informed consent to participate in this study was obtained from all the patients.
Statistical analysis
A database was created using Microsoft Excel (Microsoft® Excel for Windows, Microsoft, Redmond, WA, USA) and analyzed using commercial software (GraphPad Prism version 9; GraphPad Software, San Diego, CA, USA). Survival curves were constructed using the Kaplan–Meier method, and the log-rank test was used to evaluate the statistical significance of differences. All experiments were carried out in triplicate, and the data shown are representative of consistently observed results. The data are presented as the mean ± standard error of the mean (SE) values. The Mann–Whitney U test was used to compare continuous variables between the two groups. Categorical variables were compared using the χ2 test. P values less than 0.05 were considered to indicate statistically significant differences.
Results
MUC20 is highly expressed in gastric SRCC cells and correlated with a poor prognosis in GC patients
To investigate the key molecules involved in gastric SRCC-specific characteristics, we first examined gene expression by comparing the genome-wide mRNA expression profiles of GC cell lines (SRCC and non-SRCC cells) from the GEMINI (GSE22183) dataset, which we reported in a previous study [21]. Based on the comparison, the expression of 16 genes, including 14 protein-coding genes, was significantly higher in SRCC cell lines than in non-SRCC cell lines. Moreover, we screened 5 genes (KLK7, PRSS21, SPNS2, MUC20 and CLDN18) specifically expressed in SRCC cells among 14 protein-coding genes (Fig. 1A). Given that poor cohesion and mucin production are well-known characteristics of gastric SRCC cells (Supplementary Fig. 1 A), we focused on Kallikrein-related peptidase 7 (KLK7), a serine protease mediating the proteolytic degradation of adhesion proteins [22], and MUC20, a member of the mucin family. To investigate whether these two genes have an impact on the prognosis of GC patients, we first analyzed RNA sequencing (RNA-seq) data from The Cancer Genome Atlas (TCGA) using the Tumor IMmune Estimation Resource 2.0 (TIMER 2.0) database and showed that KLK7 and MUC20 mRNA expression was significantly higher in several cancer tissues, including GC tissues, than in the corresponding normal tissues (Supplementary Fig. 1 B). Moreover, the prognostic value of KLK7 and MUC20 mRNA expression in GC was investigated in Kaplan–Meier plotter online database, and the analysis indicated that high KLK7 and MUC20 mRNA expression was associated with a worse prognosis (Fig. 1B). Then, we examined the protein expression of KLK7 and MUC20 by immunohistochemistry (IHC) analysis of surgically resected tissues from 360 GC patients in the Kumamoto University cohort. Based on the protein level of MUC20 and KLK7 in GC tissues, patients were allocated into low and high groups (Fig. 1C). According to correlation analysis between the expression levels and clinicopathological features, only MUC20 protein expression was significantly correlated with the histological type of SRCC in human GC tissues (Supplementary Tables 1, 2). We subsequently assessed the relationship between GC patient survival and these two proteins by Kaplan–Meier analysis. There was no correlation between KLK7 expression and prognosis; however, GC patients with high MUC20 expression had significantly shorter overall survival than those with low MUC20 expression (p = 0.0021) (Fig. 1D). Moreover, subpopulation analysis of the cohort of stage IV GC patients revealed that GC patients with high MUC20 expression who received chemotherapy showed remarkably shorter overall survival (p = 0.0369) (Fig. 1E).
Given that MUC20 expression is significantly associated with a poor prognosis and the SRCC histological-type in GC patients, we next examined MUC20 expression at the mRNA and protein levels in GC cell lines. Consequently, we confirmed that MUC20 expression was higher in SRCC cells than in non-SRCC cells (Fig. 1F, G). Moreover, immunofluorescence (IF) staining revealed that MUC20 was specifically expressed in SRCC cells but not in non-SRCC cells (Fig. 1H).
In general, MUC20 has been reported to be a membrane-associated protein that senses the external environment and activates intracellular signal transduction pathways [23]. However, when SRCC cells were subjected to IF staining, MUC20 expression was predominantly detected in the cytoplasm of SRCC cells (Fig. 1H). To further investigate the intracellular location of MUC20, we examined MUC20 expression in SRCC cells with or without permeabilization treatment. Consequently, we confirmed that MUC20 expression was higher in permeabilized cells than in nonpermeabilized cells (Supplementary Fig. 1 C). These results suggest that MUC20 is weakly expressed on the membrane but abundant in the cytoplasm of SRCC cells. In support of the finding that stage IV GC patients with high MUC20 expression who received chemotherapy showed remarkably shorter overall survival, SRCC cells with intracellular MUC20 showed greater tolerance to cisplatin than non-SRCC cells (Fig. 1E, I). These findings suggest that MUC20 could enhance cell viability in SRCC cells, which leads to a poor prognosis in GC patients.
Silencing of MUC20 causes cell death in gastric SRCC
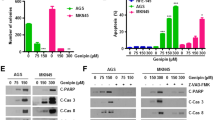
Given the correlation between MUC20 expression and a poor prognosis in GC patients, we assumed that MUC20 has an impact on the aggressive characteristics of SRCC cells. To verify its importance, we silenced MUC20 in SRCC (NUGC4 and KATOIII) cells by transfecting them with two small interfering RNAs (siRNAs). Both mRNA and protein expression were markedly decreased with siRNA treatment (Fig. 2A, B). Notably, we observed that SRCC cells transfected with MUC20 siRNAs showed cytoplasmic swelling, plasma membrane blebbing, and nuclear fragmentation (Supplementary Fig. 2). As these morphologies suggested cell death [24], we examined cell viability by IF staining with Cytotox Green Dye and confirmed the increased number of dead cells in both SRCC cell lines transfected with MUC20 siRNAs (Fig. 2C). To further explore the cell death process mediated by MUC20 silencing, we performed flow cytometry analysis in SRCC cells transfected with control siRNA or MUC20 siRNAs and demonstrated that silencing MUC20 caused programmed cell death in SRCC cells (Fig. 2D). In general, programmed cell death can occur in an active manner with specific signaling events. Several different activated signaling pathways, including apoptosis, pyroptosis and ferroptosis, have emerged as important in both normal and pathological conditions [25]. Apoptosis is widely known for caspase-3- and poly-ADP ribose polymerase (PARP)-mediated signaling pathways [26]. Cleavage of gasdermins to the gasdermin-N domain alone can signal cell pyroptosis [27]. Ferroptosis is a unique form of cell death that is driven by the accumulation of lipid-based reactive oxygen species (ROS) [28]. We first examined apoptotic signaling by Western blotting and showed that caspase-3 expression was reduced and cleaved-caspase-3 expression was increased in SRCC cells transfected with MUC20 siRNAs compared to those transfected with control siRNA (Fig. 2E). To further distinguish between apoptosis and the other cell death pathways, we also confirmed that cleaved PARP expression was increased in MUC20 knockdown SRCC cells (Fig. 2E). These results indicated that apoptosis occurred in MUC20-silenced SRCC cells. In addition to its role in the apoptotic pathway, caspase-3 can also induce pyroptosis through the cleavage of gasdermin E (GSDME) to the gasdermin-N domain [29,30,31]. Therefore, we investigated the expression of GSDME and GSDME-N fragments by Western blotting. The GSDME-N fragment was significantly increased in the MUC20-silenced groups, suggesting that MUC20 knockdown could cause pyroptosis concomitant with apoptosis in SRCC cells (Fig. 2E). In addition, a previous study showed that cytosolic lncRNA P53RRA bound Ras GTPase-activating protein-binding protein 1 and displaced p53, which could lead to apoptosis and ferroptosis [32]; thus, we investigated the lipid ROS level in MUC20-silenced SRCC cells. However, lipid ROS levels were not increased in MUC20-silenced SRCC cells (Fig. 2F), suggesting that ferroptosis is not involved in cell death induced by MUC20 silencing. Taken together, these findings indicate that MUC20 silencing could activate the apoptotic and pyroptotic programs in gastric SRCC cells, implying that cytoplasmic MUC20 may play a crucial role in the survival of gastric SRCC cells.
Silencing of MUC20 causes SRCC cell death. A Measurement of MUC20 expression in NUGC4 and KATOIII cells for 24 h after transfection with MUC20 siRNAs (#1 or #2) and control siRNA by qRT-PCR. B Western blot analysis of MUC20 in NUGC4 and KATOIII cells for 48 h after transfection with MUC20 siRNAs (#1 or #2) and control siRNA. β-Actin was similarly analyzed as a loading control. C Cytotox green dye staining of NUGC4 and KATOIII cells for 72 h after treatment with MUC20 siRNAs (#1 or #2) and control siRNA. The graph shows the percentage of Cytotox green dye-positive staining area with respect to the total area. D NUGC4 and KATOIII cells were costained with 7-AAD and annexin V for the specific identification of dead cells for 24 h after transfection with MUC20 siRNAs (#1 or #2) and control siRNA and then analyzed by flow cytometry. E Western blot analysis of caspase-3, cleaved caspase-3, cleaved PARP, GSDME full length (GSDME), GSDME-N-terminal (GSDME-N) and GSDME-C-terminal (GSDME-C) expression in NUGC4 and KATOIII cells for 48 h after transfection with MUC20 siRNAs (#1 or #2) and control siRNA. β-Actin was similarly analyzed as a loading control. F Lipid peroxidation was measured by C11-BODIPY 581/591. Histograms show the fluorescence intensity of C11-BODIPY 581/591 in NUGC4 and KATOIII cells 48 h after transfection with MUC20 siRNAs (#1 or #2) and control siRNA. (*p < 0.05, **p < 0.01, ***p < 0.005; n = 3 (A, D); n = 6 (C); scale bars, 200 μm)
Forced MUC20 expression enhanced drug resistance in GC cells
To next address the significance of MUC20 expression in GC cells, we conducted MUC20 forced expression in GC cells with low expression level of MUC20. Several kinds of MUC20 isoforms have been reported in the public Gene database (NCBI); therefore, we first investigated the variants of MUC20 and identified MUC20 variant 2 (MUC20v2), also known as isoform b, which was specifically expressed in SRCC cells (Supplementary Fig. 3 A, B). Then, we constructed an MUC20v2 expression vector and confirmed the expression of MUC20 in GC cells with either the control or MUC20v2 vector (Fig. 3A and B). Consistent with the results for SRCC cells, IF staining revealed that MUC20v2 expression was predominantly detected in the cytoplasm of MUC20v2-overexpressing cells (NUGC3-MUC20v2-OE, MKN45-MUC20v2-OE) (Fig. 3C). To next investigate the significance of MUC20v2 in GC cells, we first compared the cell proliferation and viability between the control group cells (NUGC3-MOCK, MKN45-MOCK) and MUC20v2-OE cells. However, forced MUC20v2 expression did not have an impact on cell growth or viability in NUGC3 and MKN45 cells (Fig. 3D, E). These findings suggest that these GC cells do not depend on MUC20v2 expression to survive and proliferate without additional stress. Therefore, we predicted that MUC20v2 may be involved in the survival of these GC cells under stressed circumstances. Therefore, we examined the sensitivity to cisplatin (CDDP) and paclitaxel (PTX) in control group cells and MUC20v2-OE cells; notably, both NUGC3 and MKN45 showed higher cell viability with MUC20v2 overexpression than control group cells (Fig. 3F and G). These findings indicate that MUC20v2 could enhance the viability of GC cells exposed to severe stresses, such as anticancer drug treatment.
MUC20v2 overexpression in GC cells induces chemoresistance. A Measurement of MUC20v2 expression in NUGC3 and MKN45 cells transfected with control vector and MUC20v2 overexpression vector. B Western blot analysis of MUC20v2 in NUGC3 and MKN45 cells transfected with control vector and MUC20v2 overexpression vector. The expression of β-actin was similarly analyzed as a loading control. C IF staining for MUC20v2 and wheat germ agglutinin combined with DAPI staining. D Growth assay for NUGC3 and MKN45 cells transfected with control vector and MUC20v2 overexpression vector. E Cytotox green dye staining for NUGC3 and MKN45 cells transfected with control vector and MUC20v2 overexpression vector. (F-G) Viability of NUGC3 and MKN45 cells transfected with control vector and MUC20v2 overexpression vector with cisplatin (5 µg/ml) or paclitaxel (6 µg/ml) treatment for 24 h and then cultured with normal medium. (*p < 0.05, **p < 0.01, ***p < 0.005, n.s not significant, n = 3 (A, D, F, G), n = 5 (E), scale bars, C, 25 μm; E, 200 μm)
MUC20v2 promoted cell viability by maintaining mitochondrial calcium homeostasis
To further address the mechanism of chemoresistance in MUC20v2-OE cells, we conducted RNA-seq to identify the critical pathway mediating chemoresistance. Functional and pathway enrichment analysis using 2894 DEGs revealed that the pathway involved in regulation of cytosolic calcium ion level was identified as an upregulated pathway in MUC20v2-OE NUGC3 cells, and the pathway involved in positive regulation of cytosolic calcium ion level was determined as a top of upregulated pathways by the same analysis using 2803 DEGs in MUC20v2-OE MKN45 cells (Fig. 4A). Notably, we found that genes related to calcium outflow, such as LRP1, ANXA6 and HSP90B1, were upregulated, whereas genes related to calcium internal flow, such as GRIN2A, ATP2C2 and P2RC5, were downregulated in both MUC20v2-OE cell lines (Fig. 4B). Based on the results from these analyses, we predicted that MUC20v2 may be involved in intracellular calcium ion regulation while enhancing cell survival under anticancer drug treatment conditions. Supporting a previous finding that CDDP could increase calcium level and induce cell death [33], we showed that intracellular calcium levels were increased by CDDP treatment (Supplementary Fig. 4A). In addition, we investigated the intracellular and mitochondrial calcium levels in MUC20v2-OE cells and compared them to those in MOCK cells with or without CDDP treatment. Although there was no difference in calcium concentration between MOCK cells and MUC20v2-OE cells without CDDP treatment, both cytoplastic and mitochondrial calcium levels were decreased in MUC20v2-OE cells compared with MOCK cells treated with CDDP (Fig. 4C and Supplementary Fig. 4B). Given that calcium can affect cell survival and chemoresistance in target cells [34], our present findings suggest that MUC20v2-OE GC cells show greater chemoresistance than control cells and that this is mediated by the regulation of calcium ions. Moreover, the excess accumulation of mitochondrial calcium could enhance the depolarization of the mitochondrial membrane (the loss of mitochondrial membrane potential [MMP]), which is a preliminary step toward cell death [35]. To precisely verify this theory, we performed flow cytometry to examine mitochondrial calcium levels in control and MUC20v2-OE cells treated with CDDP or PTX. Consequently, the number of cells with high mitochondrial calcium was lower in the MUC20v2-OE groups than in the control groups with anticancer drug treatment (Fig. 4D). Next, JC-1 dye was used to detect the MMP of MUC20v2-OE cells with anticancer drugs. JC-1 red is known as an indicator for MMP and JC-1 green reflects mitochondrial mass. According to the results for JC-1 staining, MMP was maintained in MUC20v2-OE cells but not in control cells after anticancer drug treatment (Fig. 4E). These findings suggest that MUC20v2 has a crucial role in maintaining mitochondrial calcium homeostasis under anticancer drug treatment.
MUC20v2 enhances chemoresistance by regulating mitochondrial calcium homeostasis. A Functional and pathway enrichment analysis of RNA-seq data for NUGC3 and MKN45 cells transfected with control vector and MUC20v2 overexpression vector. B Heatmap showing specific genes that regulate calcium homeostasis, which were expressed at higher and lower levels in MUC20v2-OE cells (NUGC3 and MKN45) than in MOCK cells (NUGC3 and MKN45). C After culture of the MOCK cells (NUGC3 and MKN45) and MUC20v2-OE cells (NUGC3 and MKN45) under different conditions (normal medium and 5 µg/ml CDDP) for 18 h, intracellular Ca2 + and mitochondrial Ca2 + levels were examined by IF staining with Fluo-4 and Rhod-2. The graph shows the percentage of red- and green-positive cells. D After culture of MOCK cells (NUGC3 and MKN45) and MUC20v2-overexpressing cells (NUGC3 and MKN45) under different conditions (5 µg/ml CDDP or 6 µg/ml PTX) for 24 h, mitochondrial Ca2 + levels were examined by Rhod-2 staining. E After culture of MOCK cells (NUGC3 and MKN45) and MUC20v2-OE cells (NUGC3 and MKN45) under different conditions (5 µg/ml CDDP or 6 µg/ml PTX) for 24 h, MMP was examined by JC-1 staining. The graph shows the proportion of JC-1-red and JC-1-green staining. F Western blot analysis of MUC20, caspase-3, cleaved caspase-3, cleaved PARP, GSDME, GSDME-N and GSDME-C expression in MOCK cells (NUGC3 and MKN45) and MUC20v2-OE cells (NUGC3 and MKN45) subjected to different conditions (5 µg/ml CDDP or 6 µg/ml PTX) for 24 h. β-Actin was similarly analyzed as a loading control. G MOCK cells (NUGC3 and MKN45) and MUC20v2-OE cells (NUGC3 and MKN45) were costained with 7-AAD and annexin V for the specific identification of dead cells for 24 h after treatment with anticancer drugs (5 µg/ml CDDP) and then analyzed by flow cytometry. The graph shows the percentage of dead cells. (*p < 0.05, **p < 0.01, n.s not significant, n = 3; scale bars, 25 μm)
Moreover, we examined the apoptotic and pyroptotic pathways and programmed cell death in MUC20v2-OE cells treated with anticancer drugs. Cleaved caspase-3, cleaved PARP, and GSDME-N expression and the number of dead cells were decreased in MUC20v2-OE cells compared with control cells after anticancer drug treatment (Fig. 4 F, G), suggesting that programmed cell death induced by anticancer drugs is inhibited by forced MUC20v2 expression in GC cells.
MUC20 silencing in SRCC cells caused abnormal calcium accumulation in mitochondria and subsequent depolarization of the mitochondrial membrane
Given that MUC20v2 overexpression could induce chemoresistance in GC cells through a reduction in calcium inside mitochondria, we next investigated whether the calcium concentration could be increased by MUC20 silencing in SRCC cells. We first analyzed cytoplastic and mitochondrial calcium levels by IF staining. Although the cytoplastic calcium levels were not remarkably changed in SRCC cells transfected with MUC20 siRNAs, the mitochondrial calcium levels were significantly increased in SRCC cells transfected with MUC20 siRNAs compared with those transfected with control siRNA (Fig. 5A). Flow cytometric analysis also revealed that mitochondrial calcium levels were increased by MUC20 silencing with both siRNAs (Fig. 5B). Moreover, MMP evaluated by JC-1 dye was reduced by MUC20 silencing in SRCC cells (Fig. 5C). Combined with the results regarding MUC20 silencing-induced programmed cell death (Fig. 2D, E), these findings suggest that MUC20v2 in SRCC cells suppresses apoptosis and pyroptosis by regulating mitochondrial calcium levels.
MUC20 silencing causes mitochondrial calcium overload and subsequent depolarization of the mitochondrial membrane. A IF staining with Fluo-4 and Rhod-2 for NUGC4 and KATOIII cells after transfection with MUC20 siRNAs (#1 or #2) and control siRNA. The graph shows the percentage of red-positive cells. B NUGC4 and KATOIII cells were stained with Rhod-2 after transfection with MUC20 siRNAs (#1 or #2) and control siRNA and then analyzed by flow cytometry. C Staining with JC-1 in NUGC4 and KATOIII cells after transfection with MUC20 siRNAs (#1 or #2) and control siRNA and then analysis by flow cytometry. The graph shows the proportion of JC-1-red and JC-1-green staining. (*p < 0.05, **p < 0.01; n = 3; scale bars, 25 μm)
Forced MUC20v2 expression in GC cells markedly enhanced chemoresistance in a xenograft mouse model
To verify the significance of MUC20v2 in GC chemoresistance in an in vivo model, NUGC3-MOCK cells and NUGC3-MUC20v2-OE cells were injected subcutaneously into nude mice. The mice were subsequently treated with PBS and CDDP according to the protocol shown in Fig. 6A. CDDP treatment significantly reduced the size of subcutaneous tumors derived from MOCK cells, whereas treatment with CDDP did not exert an antitumor effect in the tumors derived from MUC20v2-OE cells (Fig. 6B, C), which suggests that MUC20v2 plays a role in the chemoresistance of GC cells in vivo as well as in vitro. In addition, TUNEL staining showed that apoptotic tumor cells were significantly increased in the tumors derived from MOCK cells compared to those of MUC20v2-OE cells under CDDP treatment (Fig. 6D). These results suggest that MUC20v2 suppresses GC cell death induced by anticancer drugs through maintaining mitochondrial calcium homeostasis in vivo.
MUC20v2 overexpression in GC cells markedly enhances chemoresistance in an in vivo model. A MOCK cells and MUC20v2-OE cells orthotopic xenografts were injected subcutaneously into mice. After 21 days, the mice were injected intraperitoneally with PBS (MOCK and MUC20) or cisplatin (MOCK and MUC20) once every 2 days. 2 days after injection, the mice were euthanized, and the tumors were dissected and weighed. B Images showing tumor size. C Statistical analysis of tumor size and weight. D Representative hematoxylin and eosin (H, E) and TUNEL staining of tumors from mice and quantification of MUC20-positive and TUNEL-positive cells. E Proposed model of the relationship between intracellular MUC20 expression and mitochondrial calcium homeostasis in GC cancer cells. (*p < 0.05, n.s not significant, n = 8; scale bars, 50 μm.)
Discussion
SRCC is known for its specific histological phenotype among all WHO classification types in GC, including unique cellular morphology and mucin-producing characteristics, and it also has a worse prognosis and lower chemosensitivity than the other types [9, 10]. In the current study, we tried to investigate the molecular mechanism related to these aggressive characteristics in SRCC. According to the genome-wide mRNA expression profiles of GC cell lines, we found 16 genes that were specifically highly expressed in gastric SRCC cells. Based on the histological characteristics of SRCC and survival analysis, we narrowed them down to MUC20, which is a member of the mucin family. Mucins are high molecular weight glycoproteins that play a role in physiological protection and lubrication to epithelial surfaces. Due to the presence or absence of the transmembrane domain, mucins are classified into two categories: secreted and membrane-bound types [36]. Between these two types, the membrane-bound type is in the spotlight for its alterations in the growth and survival of cancer cells [37]. MUC20 is a membrane-bound mucin that has been reported to be correlated with the progression of various cancers, such as thyroid cancer, ovarian cancer, colorectal cancer, pancreatic cancer and endometrial cancer [38,39,40,41,42]. In addition, a pancancer study based on TCGA transcriptome data indicated that the overexpression of MUC20 in GC predicted a poor prognosis [43]. In the present study, we demonstrated that MUC20 expression had a poor prognostic impact on chemotherapy-treated GC patients as well as a cohort of 360 GC patients. However, further investigation in a large SRCC patient cohort will be required to prove the significance of MUC20 expression among gastric SRCC patients.
Previous evidence showed that the interaction of MUC20-MET and MUC4-HER2 at the cell membrane could regulate downstream cell signaling events [36]. Overexpressed MUC20 activates the EGFR-STAT3 pathway, promotes epidermal growth factor receptor (EGF) expression, and induces a malignant phenotype of endometrial cancer [42]. In addition, MUC20 enhances the expression of hepatocyte growth factor (HGF), a ligand of MET, and then promotes HGF-dependent MET and AKT phosphorylation in pancreatic cancer [41]. Although we first found that MUC20v2 is predominantly expressed in the cytoplasm and not the membrane of gastric SRCC cells, the intracellular MUC20v2-mediated signaling pathway is unknown. Here, we showed that the expression of calcium efflux-associated genes increased while that of calcium influx-related genes decreased in MUC20v2-overexpressing GC cells. In general, calcium overload in mitochondria can activate the caspase-3-mediated apoptotic pathway [44]. Supporting this theory, we demonstrated that MUC20v2 overexpression inhibits the accumulation of calcium in mitochondria to protect GC cells from chemotherapy-induced death. Moreover, we showed that calcium was overloaded in mitochondria in MUC20-silenced SRCC cells, which resulted in cell death.
Recent evidence shows that chemotherapy can induce pyroptosis via the cleavage of GSDME by activated caspase-3 to generate the GSDME-N fragment in cancer cells [29,30,31]. Our findings also showed a subsequent increase in the levels of the GSDME-N fragment and cleaved PARP after knockdown of MUC20v2. This shows that deficiency of MUC20v2 could promote cell death through the apoptosis and pyroptosis forms of programmed cell death. Mitochondria are known to play many roles in cancer cells, including development, stress response, wound healing and nutritional status [45]. MMP is a universal selective indicator of mitochondrial function, and alterations in MMP are involved in many human pathologies [46]. For instance, high MMP is a major marker for mitochondrial activity and cell health; in contrast, low MMP is related to mitochondrial dysfunction and cell death [47, 48]. Consistent with these theories, MUC20v2 knockdown causes mitochondrial calcium overload and subsequent MMP depolarization in SRCC cells. Moreover, when MUC20v2-OE GC cells were treated with anticancer drugs, MUC20v2 maintained the mitochondrial calcium concentration and MMP to evade cell death. Generally, MMP alteration is associated with lipid ROS accumulation and ferroptosis, and inhibition of the TCA cycle alleviates MMP hyperpolarization to reduce ferroptosis [28]. However, we showed that MUC20v2 knockdown led to MMP depolarization and decreased the lipid ROS level in SRCC cell cells. This finding suggests that programmed cell death caused by MUC20v2 knockdown is not correlated with ferroptosis.
In conclusion, MUC20v2 is dominantly expressed in the cytoplasm of gastric SRCC cells. MUC20v2 enhances cell survival ability and chemoresistance by maintaining mitochondrial calcium levels and MMP to protect GC cells from apoptosis and pyroptosis. In addition, MUC20 silencing in SRCC cells causes calcium overload in mitochondria and subsequent depolarization of the mitochondrial membrane, which leads to caspase-3-mediated programmed cell death (Fig. 6 E). Therefore, MUC20v2 could be a novel therapeutic target for eliminating SRCC cells and improving the prognosis of gastric SRCC patients.
Abbreviations
- SRCC:
-
Signet ring cell carcinoma
- GC:
-
Gastric cancer
- MUC20:
-
Mucin 20
- KLK7:
-
Kallikrein-related peptidase 7
- MUC20v2:
-
MUC20 variant 2
- PARP:
-
Poly-ADP ribose polymerase
- GSDME:
-
Gasdermin
- Lipid ROS:
-
Lipid-based reactive oxygen species
- CASP3:
-
Caspase3
- OE:
-
Overexpression
References
Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Chen YC, Fang WL, Wang RF, et al. Clinicopathological variation of lauren classification in gastric cancer. Pathol Oncol Res. 2016;22:197–202.
Cimerman M, Repse S, Jelenc F, et al. Comparison of Lauren’s, Ming’s and WHO histological classifications of gastric cancer as a prognostic factor for operated patients. Int Surg. 1994;79:27–32.
Lai JF, Xu WN, Noh SH, et al. Effect of world health organization (WHO) histological classification on predicting lymph node metastasis and recurrence in early gastric cancer. Med Sci Monit. 2016;22:3147–53.
Liu X, Cai H, Sheng W, et al. Clinicopathological characteristics and survival outcomes of primary signet ring cell carcinoma in the stomach: retrospective analysis of single center database. PLoS ONE. 2015;10:e0144420.
Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9.
Mariette C, Carneiro F, Grabsch HI, et al. Consensus on the pathological definition and classification of poorly cohesive gastric carcinoma. Gastric Cancer. 2019;22:1–9.
Pernot S, Voron T, Perkins G, et al. Signet-ring cell carcinoma of the stomach: impact on prognosis and specific therapeutic challenge. World J Gastroenterol. 2015;21:11428–38.
Bamboat ZM, Tang LH, Vinuela E, et al. Stage-stratified prognosis of signet ring cell histology in patients undergoing curative resection for gastric adenocarcinoma. Ann Surg Oncol. 2014;21:1678–85.
Taghavi S, Jayarajan SN, Davey A, et al. Prognostic significance of signet ring gastric cancer. J Clin Oncol. 2012;30:3493–8.
Tsugeno Y, Nakano K, Nakajima T, et al. Histopathologic analysis of signet-ring cell carcinoma in situ in patients with hereditary diffuse gastric cancer. Am J Surg Pathol. 2020;44:1204–12.
Chu PG, Weiss LM. Immunohistochemical characterization of signet-ring cell carcinomas of the stomach, breast, and colon. Am J Clin Pathol. 2004;121:884–92.
Yamashina M. A variant of early gastric carcinoma. histologic and histochemical studies of early signet ring cell carcinomas discovered beneath preserved surface epithelium. Cancer. 1986;58:1333–9.
Kerckhoffs KGP, Liu DHW, Saragoni L, et al. Mucin expression in gastric- and gastro-oesophageal signet-ring cell cancer: results from a comprehensive literature review and a large cohort study of Caucasian and Asian gastric cancer. Gastric Cancer. 2020;23:765–79.
Shu Y, Zhang W, Hou Q, et al. Prognostic significance of frequent CLDN18-ARHGAP26/6 fusion in gastric signet-ring cell cancer. Nat Commun. 2018;9:2447.
Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509–14.
Szasz AM, Lanczky A, Nagy A, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–33.
Ishimoto T, Miyake K, Nandi T, et al. Activation of transforming growth factor beta 1 signaling in gastric cancer-associated fibroblasts increases their motility, via expression of rhomboid 5 homolog 2, and ability to induce invasiveness of gastric cancer cells. Gastroenterology. 2017;153:191–204 (e16).
Yamada T, Ohta K, Motooka Y, et al. Significance of tsukushi in lung cancer. Lung Cancer. 2019;131:104–11.
Izumi D, Ishimoto T, Miyake K, et al. CXCL12/CXCR4 activation by cancer-associated fibroblasts promotes integrin beta1 clustering and invasiveness in gastric cancer. Int J Cancer. 2016;138:1207–19.
Ooi CH, Ivanova T, Wu J, et al. Oncogenic pathway combinations predict clinical prognosis in gastric cancer. PLoS Genet. 2009;5:e1000676.
Prassas I, Eissa A, Poda G, et al. Unleashing the therapeutic potential of human kallikrein-related serine proteases. Nat Rev Drug Discov. 2015;14:183–202.
Ballester B, Milara J, Cortijo J. Mucins as a new frontier in pulmonary fibrosis J. Clin Med. 2019;8:1447.
Kroemer G, Galluzzi L, Vandenabeele P, et al. Classification of cell death: recommendations of the nomenclature committee on cell death 2009. Cell Death Differ. 2009;16:3–11.
Khan I, Yousif A, Chesnokov M, et al. A decade of cell death studies: breathing new life into necroptosis. Pharmacol Ther. 2021;220:107717.
Uchihara T, Miyake K, Yonemura A, et al. Extracellular vesicles from cancer-associated fibroblasts containing annexin A6 induces FAK-YAP activation by stabilizing beta1 integrin. Enhancing Drug Resistance Cancer Res. 2020;80:3222–35.
Frank D, Vince JE. Pyroptosis versus necroptosis: similarities, differences, and crosstalk. Cell Death Differ. 2019;26:99–114.
Yang WS, Stockwell BR. Ferroptosis: death by lipid peroxidation. Trends Cell Biol. 2016;26:165–76.
Wang Y, Gao W, Shi X, et al. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature. 2017;547:99–103.
Li Y, Yuan Y, Huang ZX, et al. GSDME-mediated pyroptosis promotes inflammation and fibrosis in obstructive nephropathy. Cell Death Differ. 2021;28:2333–50.
Jiang M, Qi L, Li L, et al. The caspase-3/GSDME signal pathway as a switch between apoptosis and pyroptosis in cancer. Cell Death Discov. 2020;6:112.
Mao C, Wang X, Liu Y, et al. A G3BP1-Interacting lncRNA promotes ferroptosis and apoptosis in cancer via nuclear sequestration of p53. Cancer Res. 2018;78:3484–96.
Al-Taweel N, Varghese E, Florea AM, et al. Cisplatin (CDDP) triggers cell death of MCF-7 cells following disruption of intracellular calcium ([Ca(2+)]i) homeostasis. J Toxicol Sci. 2014;39:765–74.
Chakraborty PK, Mustafi SB, Xiong X, et al. MICU1 drives glycolysis and chemoresistance in ovarian cancer. Nat Commun. 2017;8:14634.
Ganta KK, Mandal A, Chaubey B. Depolarization of mitochondrial membrane potential is the initial event in non-nucleoside reverse transcriptase inhibitor efavirenz induced cytotoxicity. Cell Biol Toxicol. 2017;33:69–82.
Senapati S, Das S, Batra SK. Mucin-interacting proteins: from function to therapeutics. Trends Biochem Sci. 2010;35:236–45.
Bafna S, Kaur S, Batra SK. Membrane-bound mucins: the mechanistic basis for alterations in the growth and survival of cancer cells. Oncogene. 2010;29:2893–904.
Hou S, Xie X, Zhao J, et al. Downregulation of miR-146b-3p inhibits proliferation and migration and modulates the expression and location of sodium/iodide symporter in dedifferentiated thyroid cancer by potentially targeting MUC20. Front Oncol. 2020;10:566365.
Chen CH, Shyu MK, Wang SW, et al. MUC20 promotes aggressive phenotypes of epithelial ovarian cancer cells via activation of the integrin beta1 pathway. Gynecol Oncol. 2016;140:131–7.
Xiao X, Wang L, Wei P, et al. Role of MUC20 overexpression as a predictor of recurrence and poor outcome in colorectal cancer. J Transl Med. 2013;11:151.
Chen ST, Kuo TC, Liao YY, et al. Silencing of MUC20 suppresses the malignant character of pancreatic ductal adenocarcinoma cells through inhibition of the HGF/MET pathway. Oncogene. 2018;37:6041–53.
Chen CH, Wang SW, Chen CW, et al. MUC20 overexpression predicts poor prognosis and enhances EGF-induced malignant phenotypes via activation of the EGFR-STAT3 pathway in endometrial cancer. Gynecol Oncol. 2013;128:560–7.
Jonckheere N, Van Seuningen I. Integrative analysis of the cancer genome atlas and cancer cell lines encyclopedia large-scale genomic databases: MUC4/MUC16/MUC20 signature is associated with poor survival in human carcinomas. J Transl Med. 2018;16:259.
Pinton P, Giorgi C, Siviero R, et al. Calcium and apoptosis: ER-mitochondria Ca2+ transfer in the control of apoptosis. Oncogene. 2008;27:6407–18.
Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12:685–98.
Bazhin AA, Sinisi R, De Marchi U, et al. A bioluminescent probe for longitudinal monitoring of mitochondrial membrane potential. Nat Chem Biol. 2020;16:1385–93.
Mansell E, Sigurdsson V, Deltcheva E, et al. Mitochondrial potentiation ameliorates age-related heterogeneity in hematopoietic stem cell function. Cell Stem Cell. 2021;28:241–56 (e6).
Mishiro K, Imai T, Sugitani S, et al. Diabetes mellitus aggravates hemorrhagic transformation after ischemic stroke via mitochondrial defects leading to endothelial apoptosis. PLoS ONE. 2014;9:e103818.
Acknowledgements
We thank S. Usuki (Liaison Laboratory Research Promotion Center, IMEG, Kumamoto University) for assisting with RNA sequencing. This work was supported by the Japan Society for the Promotion of Science (JSPS, KAKENHI grant nos. 20H03531, 20K09038, 20K08985, 20K08961, 21K16384 and 21K19535), by the Naito Foundation, by The Shinnihon Foundation of Advanced Medical Treatment Research, by the Inter-University Research Network for Trans-Omics Medicine Program at the Institute of Molecular Embryology and Genetics at Kumamoto University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
There are no potential conflicts of interest to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fu, L., Yonemura, A., Yasuda-Yoshihara, N. et al. Intracellular MUC20 variant 2 maintains mitochondrial calcium homeostasis and enhances drug resistance in gastric cancer. Gastric Cancer 25, 542–557 (2022). https://doi.org/10.1007/s10120-022-01283-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-022-01283-z