Abstract
Background
The aberrant expression of long noncoding RNAs (lncRNAs) is found in various types of cancers and also showed its association with the occurrence and development of gastric cancer (GC). We found lncRNA COL1A1-014 was frequently upregulated in GC.
Methods
This study investigated COL1A1-014 for its biological function at both cellular and animal levels, using MTT, flow cytometry, colony formation and transwell assays. The expression levels of COL1A1-014 and other genes were detected by RT-PCR and western blot. Luciferase reporter assay was used to detect the potential binding of miR-1273h-5p to COL1A1-014 and CXCL12.
Results
We found that COL1A1-014 was frequently upregulated in GC tissues as well as cells. COL1A1-014 increased cell proliferation, colony forming efficiency, migration ability, invasion ability, and weight and volume of grafted tumors, while reduced cell apoptosis. Overexpression of COL1A1-014 increased the mRNA expression of chemokine (CXCmotif) ligand (CXCL12) and high levels of CXCL12 and CXCR4 proteins in GC cells. The levels of miR-1273h-5p showed an inverse correlation with COL1A1-014 and CXCL12 in GC cells transfected with miR-1273h-5p. The mRNAs of wild-type COL1A1-014 and CXCL12 showed reduction in HEK293 cells transfected with miR-1273h-5p. This suggested that COL1A1-014 functions as an efficient miR-1273h-5p sponge and as a competing endogenous RNA (ceRNA) to regulate CXCL12. The proliferative activity of COL1A1-014 on GC cells was blocked by CXCL12-CXCR4 axis inhibitor AMD-3100.
Conclusions
These findings demonstrated that COL1A1-014 play an important regulatory role in GC development by functioning as a ceRNA in regulating the CXCL12/CXCR4 axis via sponging miR-1273h-5p.
Similar content being viewed by others
Introduction
Gastric cancer (GC) is one of the most common malignancies in the world and is the second most frequent cause of cancer death [1]. Although the serum carcinoembryonic antigen (CEA), carbohydrate antigen (CA) and pepsinogen (PG) are used as diagnostic markers for GC, the early detection rate is only 5% due to lack of symptoms in the early stage [2]. There are several treatment options available, such as chemotherapy, immunotherapy and targeted therapy, for the treating GC. Targeted therapeutic drugs, including trastuzumab and ramucirumab, specifically inhibit epidermal growth factor receptor 2 (EGFR-2) and vascular endothelial growth factor receptor 2 (VEGFR-2). These drugs improved the efficacy of chemotherapy and prolonged the overall survival time of patients with advanced GC. However, the prognosis of metastatic GC still remained poor [3,4,5,6]. Therefore, it is imperative to explore alternative therapeutic targets based on the pathophysiological mechanisms of GC. Previous studies have shown that the incidence of GC showed a close association with mutations and abnormal transcription of specific genes. For example, signal transduction activator 3 (STAT3), human epidermal growth factor receptor 2 (Her-2) and bone morphogenetic protein (BMP4) are highly expressed in GC. As oncogenes, these are involved in the invasion, metastasis, proliferation, apoptosis, differentiation, infiltration and drug resistance of GC and has close correlation with poor prognosis of cancer [7, 8]. On the other hand, tumor suppressors such as phosphate and tension homology deleted on chromosome ten (PTEN) and aplasia Ras homolog one (ARH) do not often express in GC [9].
Long non-coding RNAs (LncRNAs) are a class of non-coding RNAs with a length of more than 200 nt, and they have been implicated in a variety of biological processes and diseases recently [10]. LncRNA is not a by-product of RNA polymerase II transcription, and is involved in the regulation of transcription activation and other important pathophysiological processes. It regulates signal transduction pathways associated with tumor proliferation, apoptosis, metastasis, migration and drug resistance [11,12,13,14,15]. Up- or down-regulation of certain lncRNAs induces apoptosis of tumor cells, or weakens their anti-apoptotic ability [16, 17], indicating that lncRNAs act as potential therapeutic targets and biomarkers in cancer treatment.
Although tremendous advances have been made regarding the involvement of lncRNAs in tumor progression, there are still several lncRNAs with unclear regulatory mechanisms on tumors. So, understanding the role of lncRNAs in oncogenesis has great value and potential in early diagnosis, treatment and prognosis of GC. Hence, in this study, a microarray-based screening was preformed to identify lncRNAs that are differentially expressed in GC. The identified lncRNA was characterized in cells and animals to investigate its molecular mode of action in regulating the progression of GC. This in turn provides insights to discover new therapeutic targets for treating GC.
Materials and methods
Tissue specimens
GC tissues and adjacent normal tissues (located > 5 cm away from the tumor border) were obtained from 53 GC patients at the Chinese PLA General Hospital. These patients did not undergo radiotherapy or chemotherapy before undergoing surgery. The fat tissues were removed on ice and the tissues were washed with 0.9% sodium chloride, confirmed histopathologically and then stored in liquid nitrogen. The experimental protocols were approved by the Human Research Ethics Committee of Chinese PLA General Hospital. Written informed consent was obtained from all patients.
Microarrays and sequence analysis
RNA extraction and preparation of cDNA from GC and adjacent normal tissue samples were conducted as previously described [16, 17]. Affymetrix Exon Array_Gminix lncRNA-WT v1.0 (Affymetrix, Santa Clara, CA, USA) was used for expression analysis as described previously [18, 19]. The data were normalized and the differentially expressed sequences were selected based on fold-changes of ≥ 2 and p ≤ 0.05. miRNA target genes were predicted using miRanda.
Bioinformatics analysis
miRanda (https://miranda.org.uk/) and miRcode (https://www.mircode.org/) databases were used for predicting lncRNA-miRNA interaction as described previously [20, 21]. The construction of competing endogenous RNA (ceRNA) network included 3 steps: (1) selection of lncRNAs, miRNAs and mRNAs that are up- or down-regulated > 3.0 folds and with P values < 0.05; (2) identification of lncRNAs and mRNAs that are targeted by miRNAs with experimental support from miRanda software, and the intersection from the predictive value and the measured value of the chip; and (3) co-expression between lncRNAs and mRNAs. Next, the Gene Ontology (GO) and KEGG Pathway enrichment analysis, functional analysis, and real-time reverse-transcription polymerase chain reaction (RT-PCR) were performed to validate important genes.
Lentiviral transduction and transfection
To study the effects of COL1A1-014 on cell activity, cDNA sequence of COL1A1-014 was inserted into pcDNA3.1 expression vector (Invitrogen, Shanghai, China) as described previously [22]. The miRNA mimics, miRNA inhibitors and siRNAs to knockdown COL1A1-014 expression were obtained from GenePharma (Shanghai, China). For transfection of pcDNA-COL1A1-014, miRNA mimics, miRNA inhibitors, siRNAs, the GC cells (2 × 105) were seeded at a density of 2 × 105 in 6-well plates. After culturing for 24 h (reached 80% confluence), the cells were transfected with miRNA mimics, miRNA inhibitors or siRNAs at a final concentration of 25 nmol/l using Lipofectamine RNAiMAX transfection reagent (Life Technologies, USA). The above-mentioned cells were transfected with pcDNA-COL1A1-014 at a final concentration of 1 μg/μl according to the protocol recommended by the manufacturer. After transfection for 48 h, the total RNA from the harvested cells was isolated using Trizol reagent (Invitrogen, Carlsbad, CA, USA). The empty pcDNA3.1 vector and scramble sequence of miRNA mimics, miRNA inhibitors or siRNAs were used as negative controls (NC). The transfection efficiency was evaluated by quantitative real-time PCR and visualizing positive controls by confocal microscopy (Olympus FV-1000 Inverted Microscope).
MTT and flow cytometry assays
The human normal gastric epithelial cell lines (GES-1), human GC cell lines (such as BGC-823, SGC-7901, MGC-803, MKN-45 and NCI-N87) and human embryonic kidney 293 cells (HEK293) were purchased from the Cancer Institute and Hospital of Chinese Academy of Medical Science (Beijing, China). NCI-N87 and HEK293 were maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS, heat inactivated at 56°C for 30 min), 100 U/mL penicillin and 100 U/mL streptomycin at 37°C in an incubator in humidified atmosphere of 5% CO2 atmosphere. All other cell lines were grown in RPMI 1640 medium (Life Technologies, Carlsbad, CA, USA) plus 10% FBS at 37°C in a humidified atmosphere with 5% CO2.
Cell viability was evaluated using MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2-H-tetrazolium bromide] method [23]. Briefly, the transfected BGC-823 cells were treated with MTT solution (5 mg/mL) at 37 °C for 4 h and the absorbance value (OD) was measured at 490 nm using a plate reader. The cells at logarithmic growth period were digested with 0.25% trypsin, 5 × 107 cells/L were seeded in 96 wall plate (100 μL/well) and treated with CXCL12 and C-X-C chemokine receptor (CXCR) inhibitor. CXCL12 (Boster, Wuhan, China) at a final concentration of 100 ng/ml and CXCR inhibitor AMD-3100 (Sigma, USA) at a final concentration of 1 µg/ml were added to the cells and the cells were transfected with COL1A1-014 lentivirus vectors after 24 h. After culturing for another 48 h, the cells were assayed for viability using MTT method as described above.
For apoptosis assay, the transduced BGC-823 cells were treated with 200 μmol/L 5-fluorouracil (5-FU) for 24 h. The cells were then harvested and stained with Annexin V-FITC and propidium iodide (PI), and loaded onto a flow cytometer (Bection Dikinson, USA) for analysis as reported previously [24].
Colony formation and transwell assays
To determine the colony formation ability, the transduced BGC-823 cells were digested with 0.25% trypsin, followed by mixing the single cell suspension with 0.6% Noble agarose and then plated on solidified Noble agarose bottom gel. The cells were cultured for 14 days and then counted for colonies as reported [25].
Cell migration and invasion abilities were assayed using transwell methods [26]. For invasion, the membrane in the transwell chamber was coated with matrigel basement membrane matrix. The miR-1273h-5p-transfected or control vector-transfected cells were suspended at a density of 5.0 × 105/mL in DMEM containing 1.0% bovine serum albumin (BSA). For migration, 100 μL (200 μL for invasion) was added into the upper chamber of the transwell chamber, and 600 µL (700 μL for invasion) DMEM containing 20% FBS was added into the lower chamber. The cells were incubated for 24 h. At the end of the experiments, the cells on the upper side of the membrane were carefully wiped out, and the cells that migrated or invaded into the lower side of the membrane were fixed with 4% paraformaldehyde and stained with Giemsa solution. The images were then captured, and the number of cells was counted in 5 random fields for each group and presented as mean ± SD for statistical analysis.
Tumor xenograft
Special pathogen-free (SPF) male BALB/C nude mice, aged 4–6 weeks and weighing 18–20 g were purchased from the Experimental Animal Center of PLA Medical College [Permit no. SCXK (Jing) 2016-0006] and were raised in an animal center in SPF conditions. All animal experiments and animal care protocols were approved by the Institutional Animal Care and Use Committee of the Chinese PLA General Hospital. Twenty-four mice were randomly divided into 2 groups, with 12 animals in each group. For each animal, 0.2 ml (1.5 × l06) BGC-823 cells transduced with lentivirus were subcutaneously injected into the right anterior scapula to induce tumors. The animals were weighed daily and scarified 21 days later to isolate the tumors for analysis.
Luciferase reporter assay
miR-1273h-5p expression plasmid (GV268) was purchased from GeneChem Co., Ltd. (Shanghai, China). The wild-type and mutant DNA sequences of COL1A1-014 or CXCL12 were custom synthesized by GeneChem Co., Ltd. and cloned into firefly luciferase plasmid GV272 (GeneChem). HEK-293 cells after reaching 80% confluence were cultured in 24-well plates and transfected using Lipofectamine 2000 reagent (Life Technologies) according to the manufacturer’s protocol. Firefly luciferase plasmid and miR-1273h-5p expression plasmid were co-transfected with pRL-TK Renilla luciferase vector (Promega, Madison, WI, USA) for normalization. After transfection for 48 h, the luciferase activity was measured using dual-glo luciferase assay system (Promega). All experiments were performed in triplicate.
RNA extraction and RT-PCR
Total RNA was extracted using Trizol Reagent (Qiagen, USA). The RNA quantity was measured using a nanodrop (Thermofisher, USA), and the RNA purity was evaluated by A260/A280 ratio. Reverse transcription was performed using superscript III transcript kit (Thermofisher, USA). RT-PCR was performed on MyIQ PCR system (BioRad, USA). Human glyceraldehyde-3-phosphate dehydrogenase (GAPDH, Hs03929097_g1) and β-actin were used as two housekeeping genes. PCR was carried out in a total volume of 10 μl, which containing 1.5 μl of diluted and pre-amplified cDNA, 10 μl of TaqMan gene expression master mix and 1 μl of each fluorescence TaqMan probe. The cycling conditions were as follows: 50 °C for 2 min, 95 °C for 10 min followed by 40 cycles, with each one consisting of 15 s at 95 °C and 1 min at 60 °C. The samples were run in triplicate and the mean values were calculated for each case and primers used are listed in Supplementary Table S1. The data were managed using the Applied Biosystems software RQ Manager v1.2.1, internal control was using the geometric mean of the two housekeeping genes’ Ct values. The relative expression was calculated using comparative Ct method and obtained the fold change value (2−ΔΔCt) according to the previously described protocol [27].
Western blot
Protein expression was examined using Western blotting as described in the previous study [28]. Briefly, the cells after undergoing different treatments were lysed with RIPA buffer that contains protease and phosphatase inhibitors cocktail and quantitated using the BAC kit. Polyacrylamide gel electrophoresis (SDS-PAGE) was performed using 50 μg protein, transferred onto a polyvinylidene fluoride (PVDF) membrane, and then detected by proper primary and secondary antibodies before visualization with a chemiluminescence kit. The intensity of blot signals was quantitated using ultrasensitive chemiluminescence imaging system (ChemiDocXRS+).
Statistical analysis
Data from cell experiments were obtained from at least 3 independent experiments, and all data were expressed as means ± SD. Statistical comparisons between groups were evaluated using Student’s t test. P < 0.05 was considered to be statistically significant.
Results
COL1A1-014 is up-regulated in human GC tissues and GC cells
The expression of genes in 3 pairs of GC and adjacent tissues was initially analyzed using Affymetrix Exon Array_Gminix lncRNA-WT v1.0 Array. The results revealed 130 differentially expressed lncRNAs (Fig. 1a) and 584 differentially expressed mRNAs (Fig. 1b). Some of the most significant ones are listed in Supplementary Table S2. Among them, lncRNA COL1A1-014 showed up-regulation to 5.49-fold in GC tissues when compared to adjacent tissues. To verify the up-regulation levels of 8 lncRNAs (Fig. 1c–j) and 4 mRNAs (Fig. S1), the normal cell line GES-1 and 3 GC cell lines were quantified using PCR. The results showed that the expression of COL1A1-014, CAPN13, LCN2, TCHP, WSB1 and ENST00000523236 were consistent with microarray results. The expression of COL1A1-014 was significantly higher in GC cell lines, particularly in MGC-803 cells, than in normal cell line (Fig. 1g).
Analysis of differentially expressed lncRNAs among GC and tumor-adjacent normal tissues. a Cluster analysis of differentially expressed lncRNAs between GC and tumor-adjacent normal tissues was done by microarray assay. b Cluster analysis of differentially expressed mRNAs between GC and tumor-adjacent normal tissues by microarray assay. Each column represents a sample and each row represents the expression level of a gene. Green represents low expression and red represents high expression. c–j The relative expression of 8 lncRNAs in GES-1 and BGC-823, MGC-803 and SGC-7901 cell lines was determined by RT-PCR. Data are presented as means ± SD, * and ** denote P < 0.05 and 0.01, respectively, vs GES-1. GC gastric cancer tissue, NT normal tissue
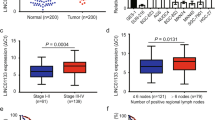
The non-coding nature of COL1A1-014 was confirmed using coding-potential analysis. There are no enough open reading frames (ORFs) for COL1A1-014, and the mutation rate of the codon on exons of COL1A1-014 are less than 0. This indicated that there is no conservative type and it is likely to be a non-coding RNA (Fig. 2a). The expression levels of COL1A1-014 were measured using RT-PCR in a total of 50 pairs of GC and adjacent tissues obtained from the GC patients (their clinical information was shown as supplementary Table S2). COL1A1-014 showed significant upregulation in 78% (39/50) of GC tissues when compared with adjacent normal tissues (Fig. 2b). The average expression level of COL1A1-014 in GC tissues was 3.47-folds when compared to the adjacent tissues in the 50 pairs of GC and adjacent tissues (Fig. 2c). Also the lymph node metastasis might be associated with the high expression of COL1A1-014 (Supplementary Table S3). The level of COL1A1-014 in GC cell lines was further measured in more cell lines. The results showed that all 5 GC cell lines (MGC-803, BGC-823, SGC-7901, MKN-45 and NCI-N87) demonstrated higher expression levels of COL1A1-014 than that of the non-tumorigenic cell line GES-1, especially MGC-803 and SGC-7901 cell lines (Fig. 2d).
The relative mRNA level of COL1A1-014 in GC tissues and cells. a Prediction of putative proteins encoded by COL1A1-014 using ORF finder, and the codon substitution frequency scores (CSF) of COL1A1-014. b, c The relative COL1A1-014 level in 50 different pairs of GC tissues and tumor-adjacent normal tissues. d The relative expression of COL1A1-014 in GES-1 and BGC-823, MGC-803, SGC-7901, MKN-45, NCI-N87 cell lines was determined by RT-PCR. Data are presented as means ± SD, * and ** denote P < 0.05 and 0.01, respectively, vs normal GC tissues or GES-1. ORF open reading frames
COL1A1-014 promoted proliferation, migration and invasion of BGC-823 cells
As COL1A1-014 showed high expression in GC tissues and cells, its impact on biological features of tumor cells was investigated. The BGC-823 cells were transduced with COL1A1-014 expression lentivirus. The lentivirus was effective to transduce BGC-823 cells with nearly 90% cells expressing GFP visual maker (Fig. 3a) and COL1A1-014 RNA level showed significant up-regulation (Fig. 3b). MTT assay showed that COL1A1-014 overexpression increased the viability of BGC-823 cells significantly within 72 h after COL1A1-014 transduction (Fig. 3c). Also COL1A1-014 decreased the apoptotic rate of BGC-823 (Fig. 3d), and even the apoptotic rate of each group was less than 5%.
Effect of COL1A1-014 on BGC-823 cell viability and apoptosis. a Transfection efficiency of COL1A1-014 under bright light and fluorescent light. b The expression of COL1A1-014 in BGC-823 cells after COL1A1-014 transfection as determined by RT-PCR. c The effect of COL1A1-014 on proliferation of BGC-823 cells at 24 h, 48 h, and 72 h as evaluated by MTT assay. d The effect of COL1A1-014 on apoptosis of BGC-823 cells for 24 h was evaluated by FCM. Data are presented as means ± SD from three independent experiments. * and ** denote P < 0.05 and 0.01, respectively, vs negative control. CON blank control, NC negative control, GFP green fluorescence protein
To further investigate the role of COL1A1-014 on cell growth and migration, soft gel assay was used to compare colony formation ability of BGC-823 cells. The results showed a significant increase in the cloning efficacy after the cells were transduced with COL1A1-014 lentivirus (Fig. 4a, b). Furthermore, the transwell assays showed that the migration (Fig. 4c, d) and invasion (Fig. 4e, f) abilities of BGC-823 cells were significantly increased after the cells were transduced with COL1A1-014 lentivirus.
Effect of COL1A1-014 on BGC-823 cells growth, invasion and migration in vitro and vivo. a, b The effect of COL1A1-014 on colony formation ability of BGC-823 cells for 24 h was evaluated by soft gel assay. c, d The effect of COL1A1-014 on migration of BGC-823 cells for 24 h was evaluated by transwell assay. e, f The effect of miR-1273h-5p on the invasion of BGC-823 cells for 24 h was evaluated by transwell assay. The effect of COL1A1-014 on tumor formation time (g), tumor formation rate (h), tumor weight (i), tumor volume (j), of BGC-823 xenograft tumors in nude mice (NC: n = 11, COL1A1-014: n = 12). k The images of mice of 21 days after transfection. Data are presented as means ± SD from 3 independent experiments. * and ** denote P < 0.05 and 0.01, respectively, vs negative control. CON blank control, NC negative control, GFP green fluorescence protein
Based on the above in vitro results, COL1A1-014 was speculated to be a sort of oncogene. To confirm this speculation, the mice were inoculated with COL1A1-014 transducted-BGC-823 cells. Observations showed that the time and rate of tumor formation between the groups were similar (Fig. 4g, h). However, COL1A1-014 significantly increased the size (volume and weight) of the tumor when compared with negative control (Fig. 4i–k).
COL1A1-014 was a target of miR-1273h-5p
To explore the possible target genes and signaling pathways involved in the tumor-promoting activity of COL1A1-014, miRanda was used to predict the potential COL1A1-014-miRNAs interactions. Three miR-1237h-5p binding sites scattering the COL1A1-014 transcripts were detected. Next, bioinformatic analysis using TargetScan and miRanda software was performed to identify the target genes for miR-1273h-5p. The results showed that CXCL12 also has a binding site in miR-1237h-5p (Fig. S2a). These 50 specimens of GC tissue were divided into COL1A1-014 high expression group (above the mean level of COL1A1-014 expression, n = 32) and low expression group (below the mean level of COL1A1-014 expression, n = 18). Interestingly, the CXCL12 level of COL1A1-014 low expression group was also significantly lower than COL1A1-014 high expression group (Fig. S2b). This suggested that the expression of CXCL12 was positively correlated with COL1A1-014 in GC tissue.
Therefore, we speculated that COL1A1-014 might act as ceRNA and competed with CXCL12 to bind to miR-1273h-5p, ultimately inhibiting the degradation of CXCL12 by miR-1273h-5p, up-regulating the intracellular level of CXCL12, and promoting the process of GC (Fig. S2c). The enriched KEGG pathways for CXCL12 were then analyzed and the results showed 16 pathways (Fig. S2d) related to cancer, which included adherens junction, leukocyte migration, signaling pathway in cancer and cell cycle. The GO enrichment of COL1A1-014 and CXCL12 included regulation of endothelial cell apoptosis process, regulation of mitotic cell cycle embryonic, DNA damage response and cell migration (Fig. S2e).
miR-1273h-5p regulated the expression of COL1A1-014 and CXCL12
As bioinformatics analysis predicted the interaction of miR-1273h-5p with both COL1A1-014 and CXCL12, the regulatory role of miR-1273h-5p on COL1A1-014 and CXCL12 was examined by transfecting the GC cells with miR-1273h-5p inhibitor and miR-1273h-5p mimics. The results showed that the levels of COL1A1-014 and CXCL12 were significantly decreased when the cells were transfected with miR-1273h-5p mimics (Fig. 5a, b). On the other hand, miR-1273h-5p inhibitor showed opposite results (Fig. 5c, d). These data suggested that COL1A1-014 and CXCL12 might be the target genes of miR-1273h-5p. To verify the specificity of miR-1273h-5p on COL1A1-014 and CXCL12, COL1A1-014 and CXCL12 reporter plasmids with mutated microRNA reaction element (MREs), in which the complementary base was substituted with non-complementary base, were constructed (Fig. 5e). Assays with HEK293 cells showed that the levels of COL1A1-014 and CXCL12 were significantly higher when mutants COL1A1-014 and CXCL12 were used (Fig. 5f, g). This indicated that miR-1273h-5p could bind to wild-type, but not mutants COL1A1-014 and CXCL12 to degrade them.
Regulation of COL1A1-014 and CXCL12 by miR-1273h-5p. a, b The CXCL12 and COL1A1-014 mRNA levels were detected by RT-PCR in GC cells after transfection with miR-1273h-5p mimics. c, d CXCL12 and COL1A1-014 mRNA levels were detected by RT-PCR in GC cells after transfection with miR-1273h-5p inhibitor. e–g The levels of COL1A1-014 and CXCL12 were detected when the wild-type/mutant COL1A1-014 and CXCL12 were used. Data are presented as means ± SD from 3 independent experiments. * and ** denote P < 0.05 and 0.01, respectively, vs negative control. WT wild-type, MUT mutant
COL1A1-014 regulated the expression of CXCL12
To further verify the relationship between COL1A1-014 and CXCL12, the COL1A1-014 levels were down- and up-regulated using siRNA and expression vector in the 3 GC cells and then the levels of CXCL12 were measured. The results showed that the siRNA down-regulated the levels of COL1A1-014 as well as CXCL12 mRNA (Fig. 6a, b), while the expression vector showed the opposite effects (Fig. 6c), which suggested that there might be a co-expression and co-regulation relationship between COL1A1-014 and CXCL12. Furthermore, the impact of COL1A1-014 on the expression of CXCL12 and CXCR4 was examined in the xenografted tumors derived from COL1A1-014-transduced BGC-823 cells. The levels of CXCL12 and CXCR4 proteins were significantly higher in the tumors derived from COL1A1-014-transduced BGC-823 cells than those from negative controls (Fig. 6d, e).
Regulation of CXCL12/CXCR4 axis expression by COL1A1-014. a, b The mRNA levels of COL1A1-014 and CXCL12 were detected by RT-PCR in GC cells after transfection with siRNA-COL1A1-014. c The mRNA levels of CXCL12 were detected by RT-PCR in GC cells after transfection with pCDNA-COL1A1-014. d The protein expression of CXCL12 and CXCR4 in xenografted tumors derived from COL1A1-014-transduced BGC-823 cells. e The representative western blots of CXCL12 and CXCR4. f COL1A1-014 might participate in the process of GC through CXCL12/CXCR4 biological axis. g The effect of COL1A1-014 on proliferation of BGC-823 cells where CXCL12/CXCR4 was blocked by AMD-3100 was evaluated by MTT assay. Data are presented as means ± SD from 3 independent experiments. * and ** denote P < 0.05 and 0.01, respectively, vs negative control. CON blank control group, NC negative control group
We herein speculated that COL1A1-014 might participate in the process of GC through CXCL12/CXCR4 biological axis (Fig. 6f). To test this, the CXCL12/CXCR4 axis was blocked to investigate the effect of COL1A1-014 on cell proliferation. MTT test showed that the proliferation rate of BGC-823 cells was increased significantly after COL1A1-014 was overexpressed and exogenous CXCL12 was supplied (p < 0.01). However, when CXCL12/CXCR4 axis antagonist AMD-3100 was added, then the increase was balanced (Fig. 6g).
Discussion
Several studies have reported the participation of lncRNAs in the progression of GC and other cancers via multiple mechanisms [7, 8]. For example, tetrandrine inhibits Wnt/β-catenin signal transduction pathways by the upregulation of Let-7a miRNA expression, and downregulation of miR-21 and miR-372 levels to arrest the GC cells at G0/G1 phase and then induce apoptosis [8]. These studies suggest that these lncRNAs might be used as markers for diagnosing GC and as targets for treating GC.
In this study, lncRNAs and mRNAs were screened based on their expression in human GC and adjacent normal tissues and also a number of differentially expressed genes have been identified. Bioinformatics analysis revealed that these genes are mainly involved in intracellular signal transduction, regulation of cell proliferation and apoptosis, and induction of chemotaxis. They are also involved in a number of signaling pathways that are associated with tumor development, such as cell migration, adhesion, cell cycle and transcriptional disorders. In particular, lncRNA COL1A1-014 is highly expressed in GC tissues and cells. COL1A1-014 (Ensemble database ID ENST00000504289.1) originates from type I collagen gene COL1A1 (Ensemble database ID ENST00000108821), and is 481 bp long. It does not encode any protein. The COL1A1 gene has 13 transcripts (splice variants), and COL1A1-014 is one of them. As it is highly expressed in GC tissues as well as cells, we speculated that it is likely related to the progression of GC.
To demonstrate this, lncRNA COL1A1-014 in BGC-823 cells was overexpressed and several assays were conducted to reveal its biological features. The results showed that lncRNA COL1A1-014 promoted growth of BGC-823 cells, suggesting that it is proliferative in tumor cells. Tumor cells often have strong ability against apoptosis induced due to chemotherapy drugs and radiotherapy [29, 30]. When BGC-823 cells were treated with 5-FU, which is a common chemotherapy drug, apoptosis was greatly increased. However, in lncRNA COL1A1-014-transfected BGC-823 cells, apoptosis was significantly lowered, which implied that COL1A1-014 has anti-apoptotic activity in GC cells. The BGC-823 cells that overexpress COL1A1-014 also showed increased ability of colony formation, which may in turn reflect the malignancy and is of great importance to prognosis [31]. It also increases the migration and invasion abilities, which are regarded as indicators of tumor metastasis [32, 33]. In addition to in vitro studies, COL1A1-014 overexpressing BGC-823 cells generated earlier and bigger tumors in mice was also found, further confirming that COL1A1-014 promotes cancer development.
LncRNA regulates mRNA to play a biological role mainly via miRNA regulation. It might competitively bind to 3′-UTR of the target gene to suppress the negative regulation by miRNA, or generate mature miRNA as a miRNA precursor to regulate the expression of the target gene. Also it may act as an endogenous miRNA sponge to compete with mRNA for MREs to inhibit the negative regulation of miRNA of the target gene, which is termed as ceRNA mechanism [34, 35]. Several studies have confirmed the existence of ceRNA regulatory mechanism in GC. For example, abnormally expressed lncRNAs such as GAPLINC, HOTAIR, BC032469, FER1L4 and MEG3 in GC competitively bind to the target genes such as CD44, HER2, hTERT, PTEN and Bcl-2 for miRNAs, causing an impact to the development and prognosis of GC [36,37,38,39,40,41].
Our analysis showed that in GC tissues with high COL1A1-014 expression, the expression level of chemokine CXCL12 showed a significant increase. Bioinformatics analysis demonstrated that the two genes have complement sequences of miR-1273h-5p. The fact that the expression of the two genes was negatively correlated with miRNA suggests that COL1A1-014 and CXCL12 might be the target genes of miR-1273h-5p. To further clearly demonstrate this, the levels of miR-1273h-5p in BGC-823 cells were up- and down-regulated with miR-1273h-5p mimics and inhibitor. The results showed that the levels of COL1A1-014 and CXCL12 are regulated by miR-1273h-5p. To further reveal the specificity of interaction, a reporter construct where the MRE was mutated with non-complemental bases was used, and found that the mutant miR-1273h-5p did not reduce the levels of COL1A1-014 and CXCL12. These data demonstrated that COL1A1-014 and CXCL12 are the targets of miR-1273h-5p. In addition, as COL1A1-014 was knocked down, the level of CXCL12 was also reduced, further proving that they are competing RNAs.
Next, the impact of COL1A1-014 on viability of GC cells with suppressed CXCL12-CXCR4 axis was investigated. The results showed that the increased cell viability by COL1A1-014 was balanced when the GC cells were treated with CXCL12-CXCR4 axis inhibitor AMD-3100. This suggested that COL1A1-014 is an oncogene in GC BGC-823 cells and might competitively bind with miR-1273h-5p to assist the inhibition of CXCL12 by miR-1273h-5p, promoting the function of CXCL12/CXCR4 axis and the proliferation and development of GC cells.
CXCL12, also known as stromal cell-derived factor-1 (SDF-1), is a member of CXC chemotactic factor subfamily. It is widely expressed in brain, lung, colon, heart and liver, and is mainly involved in the formation of blood vessels, bone marrow and lymphocytes [42,43,44]. In the last decade, chemokines and their respective receptors were well studied regarding their roles in tumor biology. The occurrence and metastasis are closely related to these factors in GC, colorectal cancer, breast cancer and melanoma [45,46,47]. CXCL12, which is the main ligand of CXCR4, is shown to stimulate the secretion of epidermal growth factor receptor (EGFR) ligands, resulting in increased migratory ability of GC cells. CXCL12/CXCR4 signaling pathway has been originally reported to regulate B lymphocyte chemoattractant [48], and then shown to play an important role in the survival, proliferation, and migration of tumor cells [49]. The binding of CXCL12 and CXCR4 to tumor cells causes activation of downstream signaling pathways, including MAPK/ERK, PI3K/Akt/NF-κB and c-Jun N terminal kinase, for regulating tumor progression [50].
Conclusions
Through microarray analysis, a number of differentially expressed lncRNAs, miRNAs and mRNAs have been identified in GC tissues. Among them, lncRNA COL1A1-014 is shown to be highly expressed in GC tissues, and promoted growth, colony formation and migratory ability of GC cells and development of grafted tumors in mice. Both COL1A1-014 and CXCL12 are considered as target genes of miR-1273h-5p and COL1A1-014 is a ceRNA of CXCL12. COL1A1-014 impacts the progression of GC by regulating CXCL12-CXCR4 axis.
References
Hartgrink HH, Jansen EP, van Grieken NC, van de Velde CJ. Gastric cancer. Lancet. 2009;374(9688):477–90.
Hamashima C. Current issues and future perspectives of gastric cancer screening. World J Gastroenterol. 2014;20:13767–74.
Goel G, Sun W. Advances in the management of gastrointestinal cancers–an upcoming role of immune checkpoint blockade. J Hematol Oncol. 2015;8:86.
Yu J, Zhang Y, Leung LH, Liu L, Yang F, Yao X. Efficacy and safety of angiogenesis inhibitors in advanced gastric cancer: a systematic review and meta-analysis. J Hematol Oncol. 2016;9:111.
Wang X, Wang C, Zhang X, Hua R, Gan L, Huang M, et al. Bmi-1 regulates stem cell-like properties of gastric cancer cells via modulating miRNAs. J Hematol Oncol. 2016;9:90.
Ansell SM, Lesokhin AM, Borrello I, Halwani A, Scott EC, Gutierrez M, et al. PD-1 blockade with nivolumab in relapsed or refractory Hodgkin's lymphoma. N Engl J Med. 2015;372:311–9.
Takehana T, Kunitomo K, Kono K, Kitahara F, Iizuka H, Matsumoto Y, et al. Status of c-erbB-2 in gastric adenocarcinoma: a comparative study of immunohistochemistry, fluorescence in situ hybridization and enzyme-linked immuno-sorbent assay. Int J Cancer. 2002;98:833–7.
Pan YM, Wang CG, Zhu M, Xing R, Cui JT, Li WM, et al. STAT3 signaling drives EZH2 transcriptional activation and mediates poor prognosis in gastric cancer. Mol Cancer. 2016;15:79.
Tang HL, Hu YQ, Qin XP, Jazag A, Yang H, Yang YX, et al. Aplasia ras homolog member I is downregulated in gastric cancer and silencing its expression promotes cell growth in vitro. J Gastroenterol Hepatol. 2012;27:1395–404.
Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136(4):629–41.
Gu Y, Chen T, Li G, Yu X, Lu Y, Wang H, et al. LncRNAs: emerging biomarkers in gastric cancer. Future Oncol. 2015;11:2427–41.
Wang Y, Zhang D, Wu K, Zhao Q, Nie Y, Fan D. Long noncoding RNA MRUL promotes ABCB1 expression in multidrug-resistant gastric cancer cell sublines. Mol Cell Biol. 2014;34:3182–93.
Qi Y, Ooi HS, Wu J, Chen J, Zhang X, Tan S, et al. MALAT1 long ncRNA promotes gastric cancer metastasis by suppressing PCDH10. Oncotarget. 2016;7:12693–703.
Qiu M, Xu Y, Yang X, Wang J, Hu J, Xu L, et al. CCAT2 is a lung adenocarcinoma-specific long non-coding RNA and promotes invasion of non-small cell lung cancer. Tumour Biol. 2014;35:5375–80.
Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J, et al. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159–65.
Trimarchi T, Bilal E, Ntziachristos P, Fabbri G, Dalla-Favera R, Tsirigos A, et al. Genome-wide mapping and characterization of Notch-regulated long noncoding RNAs in acute leukemia. Cell. 2014;158:593–606.
Rossi MN, Antonangeli F. LncRNAs: New Players in Apoptosis Control. Int J Cell Biol. 2014;2014:473857.
Noriega NC, Eghlidi DH, Garyfallou VT, Kohama SG, Kryger SG, Urbanski HF. Influence of 17beta-estradiol and progesterone on GABAergic gene expression in the arcuate nucleus, amygdala and hippocampus of the rhesus macaque. Brain Res. 2010;1307:28–422.
Noriega NC, Garyfallou VT, Kohama SG, Urbanski HF. Glutamate receptor subunit expression in the rhesus macaque locus coeruleus. Brain Res. 2007;1173:53–655.
Oulas A, Karathanasis N, Louloupi A, Pavlopoulos GA, Poirazi P, Kalantidis K, et al. Prediction of miRNA targets. Methods Mol Biol. 2015;1269:207–29.
Peterson SM, Thompson JA, Ufkin ML, Sathyanarayana P, Liaw L, Congdon CB. Common features of microRNA target prediction tools. Front Genet. 2014;5:23.
Savan R, Chan T, Young HA. Lentiviral gene transduction in human and mouse NK cell lines. Methods Mol Biol. 2010;612:209–21.
Sylvester PW. Optimization of the tetrazolium dye (MTT) colorimetric assay for cellular growth and viability. Methods Mol Biol. 2011;716:157–68.
Christensen ME, Jansen ES, Sanchez W, Waterhouse NJ. Flow cytometry based assays for the measurement of apoptosis-associated mitochondrial membrane depolarisation and cytochrome c release. Methods. 2013;61:138–45.
Borowicz S, Van Scoyk M, Avasarala S, Karuppusamy Rathinam MK, Tauler J, Bikkavilli RK, et al. The soft agar colony formation assay. J Vis Exp. 2014;92:e51998.
Liu X, Wu X. Utilizing matrigel transwell invasion assay to detect and enumerate circulating tumor cells. Methods Mol Biol. 2017;1634:277–82.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8.
Hnasko TS, Hnasko RM. The Western Blot. Methods Mol Biol. 2015;1318:87–96.
Shen DW, Pouliot LM, Hall MD, Gottesman MM. Cisplatin resistance: a cellular self-defense mechanism resulting from multiple epigenetic and genetic changes. Pharmacol Rev. 2012;64:706–21.
Botti G, Scognamiglio G, Marra L, Pizzolorusso A, Di Bonito M, De Cecio R, et al. Programmed death ligand 1 (PD-L1) expression in primary angiosarcoma. J Cancer. 2017;8:3166–72.
Wang MW, Liu J, Liu Q, Xu QH, Li TF, Jin S, et al. LncRNA SNHG7 promotes the proliferation and inhibits apoptosis of gastric cancer cells by repressing the P15 and P16 expression. Eur Rev Med Pharmacol Sci. 2017;21:4613–22.
Mathews LA, Hurt EM, Zhang X, Farrar WL. Genomic analysis of invasive human bone marrow derived mesenchymal stem cells. J Bone Marrow Res. 2013;1:122.
Shelef MA, Bennin DA, Yasmin N, Warner TF, Ludwig T, Beggs HE, et al. Focal adhesion kinase is required for synovial fibroblast invasion, but not murine inflammatory arthritis. Arthritis Res Ther. 2014;16:464.
Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, et al. LincRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47:648–55.
Wang Y, Xu Z, Jiang J, Xu C, Kang J, Xiao L, et al. Endogenous miRNA sponge lincRNA-RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self-renewal. Dev Cell. 2013;25:69–80.
Hu Y, Wang J, Qian J, Kong X, Tang J, Wang Y, et al. Long noncoding RNA GAPLINC regulates CD44-dependent cell invasiveness and associates with poor prognosis of gastric cancer. Cancer Res. 2014;74:6890–902.
Pan W, Liu L, Wei J, Ge Y, Zhang J, Chen H, et al. A functional lncRNA HOTAIR genetic variant contributes to gastric cancer susceptibility. Mol Carcinog. 2016;55:90–6.
Lu MH, Tang B, Zeng S, Hu CJ, Xie R, Wu YY, et al. Long noncoding RNA BC032469, a novel competing endogenous RNA, upregulates hTERT expression by sponging miR-1207-5p and promotes proliferation in gastric cancer. Oncogene. 2016;35:3524–34.
Xia T, Liao Q, Jiang X, Shao Y, Xiao B, Xi Y, et al. Long noncoding RNA associated-competing endogenous RNAs in gastric cancer. Sci Rep. 2014;4:6088.
Guo J, Miao Y, Xiao B, Huan R, Jiang Z, Meng D, et al. Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol. 2009;24:652–7.
Peng W, Si S, Zhang Q, Li C, Zhao F, Wang F, et al. Long non-coding RNA MEG3 functions as a competing endogenous RNA to regulate gastric cancer progression. J Exp Clin Cancer Res. 2015;34:79.
Izumi D, Ishimoto T, Miyake K, Sugihara H, Eto K, Sawayama H, et al. CXCL12/CXCR4 activation by cancer-associated fibroblasts promotes integrin beta1 clustering and invasiveness in gastric cancer. Int J Cancer. 2016;138:1207–19.
Ling KX, Peng L, Feng ZJ, Wei C, Yan YW, Nan S, et al. Stromal derived factor-1/CXCR4 axis involved in bone marrow mesenchymal stem cells recruitment to injured liver. Stem Cells Int. 2016;2016:8906945.
Satomura H, Sasaki K, Nakajima M, Yamaguchi S, Onodera S, Otsuka K, et al. Can expression of CXCL12 and CXCR4 be used to predict survival of gastric cancer patients? Anticancer Res. 2014;34:4051–7.
Guo ZJ, Yang L, Qian F, Wang YX, Yu X, Ji CD, et al. Transcription factor RUNX2 up-regulates chemokine receptor CXCR4 to promote invasive and metastatic potentials of human gastric cancer. Oncotarget. 2016;7:20999–1012.
Teng F, Tian WY, Wang YM, Zhang YF, Guo F, Zhao J, et al. Cancer-associated fibroblasts promote the progression of endometrial cancer via the SDF-1/CXCR4 axis. J Hematol Oncol. 2016;9:8.
Zhao BC, Wang ZJ, Mao WZ, Ma HC, Han JG, Zhao B, et al. CXCR4/SDF-1 axis is involved in lymph node metastasis of gastric carcinoma. World J Gastroenterol. 2011;17:2389–96.
Nagasawa T, Hirota S, Tachibana K, Takakura N, Nishikawa S, Kitamura Y, et al. Defects of B-cell lymphopoiesis and bone-marrow myelopoiesis in mice lacking the CXC chemokine PBSF/SDF-1. Nature. 1996;382:635–8.
Ratajczak MZ, Zuba-Surma E, Kucia M, Reca R, Wojakowski W, Ratajczak J. The pleiotropic effects of the SDF-1-CXCR4 axis in organogenesis, regeneration and tumorigenesis. Leukemia. 2006;20:1915–24.
Cojoc M, Peitzsch C, Trautmann F, Polishchuk L, Telegeev GD, Dubrovska A. Emerging targets in cancer management: role of the CXCL12/CXCR4 axis. Onco Targets Ther. 2013;6:1347–61.
Acknowledgements
This work was supported by the National Nature and Science Foundation of China (no. 81773778) and China Postdoctoral Science Foundation (2019M650775).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
No potential conflicts of interest were disclosed.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dong, XZ., Zhao, ZR., Hu, Y. et al. LncRNA COL1A1-014 is involved in the progression of gastric cancer via regulating CXCL12-CXCR4 axis. Gastric Cancer 23, 260–272 (2020). https://doi.org/10.1007/s10120-019-01011-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-019-01011-0