Abstract
Background
Inhibitor of growth 4 (ING4) has deserved attention as a tumor suppressor gene in many malignant tumors. In our study, we investigated ING4 immunoexpression in gastrointestinal stromal tumors (GISTs) and its prognostic value.
Method
The expression of ING4 and Ki67 was investigated in 41 samples of various risk gastrointestinal stromal tumors by immunohistochemical technique. The associations of ING4 expression and clinicopathological parameters, and prognosis of the patients, were analyzed by multivariate Cox regression analysis.
Results
ING4 expression showed a decreased trend from lower-risk to high-risk gastrointestinal stromal tumors, and an opposite trend for Ki67 expression. In lower-risk tumors, it was found the expression level of ING4 was 78.95 % ± 27.90 % and that of Ki67 was 4.42 % ± 3.75 %. However, in high-risk tumors, the expression level of ING4 was 9.23 % ± 7.66 % and that of Ki67 was 18.50 % ± 9.09 %. There was a strongly negative correlation between the expression levels of ING4 and Ki67. A significant difference was observed in the expression of ING4 between invasion and non-invasion (p < 0.001). The expression of ING4 was markedly correlated with tumor size (p < 0.001), mitotic index (p < 0.001), tumor necrosis (p = 0.021), invasion (p < 0.001), recurrence and metastasis (p = 0.021), and mortality (p < 0.001).
Conclusion
The low expression level of ING4 protein was correlated with high-risk GISTs. ING4 might be involved in the progression of GISTs and inhibit its invasion. ING4 might be one of the prognostic indicators in GISTs.
Similar content being viewed by others
Introduction
Gastrointestinal stromal tumors (GISTs) originate from the interstitial cells of Caja within the gastrointestinal tract. They are the most common type of gastrointestinal mesenchymal neoplasms, accounting for 0.1–0.3 % in gastrointestinal tract malignant tumors and 5 % in soft tissue sarcomas [1]. The incidence of GISTs is estimated to be 1–2 per 100,000 persons every year [1, 2]. GISTs occur in all ages of the population, mostly between 55 and 65 years, with a median age of 60 years old, and occur in both sexes equally [3–6]. GISTs occur mainly in the stomach (50–60 %) and small intestine (30–40 %), and also occur in the colorectum (5–10 %) and esophagus (0–5 %), with rare cases in mesentery, omentum, and retroperitoneum [1, 7]. Up to now, the prognosis of GISTs has relied clinically on tumor size, mitosis, and location, but exceptions exist. Therefore, additional prognostic indicators are still needed.
The ING (inhibitor of growth) family is a group of important tumor suppressor genes that are necessary to p53 transcriptional activity [8]. The family members including ING1, ING2, ING3, ING4, and ING5 widely participate in regulating genetic transcription, cell proliferation, apoptosis, cell senescence, contact inhibition, DNA repair, and angiogenic inhibition [8–15]. ING4, as one of the new ING family members, was discovered in 2003 [8]. It is located in chromatin 12p13.31, including 8 exons and 7 introns. The full-length cDNA is 1,380 bp, encoding 248 amino acids. ING4 is a nuclear protein expressed in normal human tissues, but its expression is markedly reduced in human gliomas [16], head and neck squamous cell carcinomas, malignant melanomas, and lung cancers [17–19], with levels inversely correlated with the tumor grades and risks [18, 20]. As a new member of the tumor suppressor genes, ING4 was thought to play important roles in malignant tumors. It was implicated in a variety of processes, such as the regulation of transcription, cell proliferation, apoptosis, cell senescence, contact inhibition, DNA repair, inhibition of tumor invasion, and metastasis [19, 21, 22].
ING4 is prone to deletion mutations and downregulation in human malignancies: its downregulation was found in various human cancer cell lines. The ING4 gene is frequently altered: in human breast cancers, the ING4 locus is frequently deleted [23]. Also, in head and neck squamous cell carcinomas, loss of heterozygosity in the ING4 gene is frequent [17]. Extrinsic expression of ING4 in a breast cancer cell line inhibits growth of the cells in soft agar [23]. It was considered to be associated with carcinogenesis and degree of malignancies [16, 17, 24]. The foregoing findings suggest that downregulation or absence of ING4 expression affects the prognosis of malignant tumors. This study was designed to investigate ING4 protein expression in GISTs by immunohistochemistry and its relationship with clinicopathological parameters and prognosis of GISTs.
Materials and methods
Patients and tumor specimens collection
Forty-one samples of formalin-fixed and paraffin-embedded tissues from 41 patients diagnosed as having gastrointestinal stromal tumors were collected from the Department of Pathology, the Third Affiliated Hospital of Harbin Medical University, between January 2005 and June 2007. The patients included 20 men and 21 women whose age ranged from 16 to 77 years, with the mean age 52.52 ± 13.21 years. These samples were prepared by hematoxylin and eosin (H&E) staining and then were diagnosed by two pathological experts. Fletcher’s criteria were adopted to predict the activity of the tumor tissues [25, 26]. Tumors were graded according to their sizes and karyokinetic states: very low risk, <2 cm and <5 mitoses/50 high-power fields (HPF); low risk, 2–5 cm and <5 mitoses/50 HPF; intermediate risk, 5–10 cm and <5 mitoses/50 HPF; high risk, >5 cm and >5 mitoses/50 HPF, >10 cm and any mitotic index, any size and >10 mitoses/50 HPF. All samples were divided into two groups: the lower-risk group (including very low, low-, and intermediate-risk GISTs) and the high-risk group (including high-risk GISTs). As shown in Table 1, the lower-risk group included 19 samples and the high-risk group included 22 samples. The clinicopathological features of all patients, including age, sex, size, necrosis, degree of risk, invasion, recurrence and/or metastasis, follow-up, and survival or death were recorded (Table 2).
Immunohistochemistry
Immunohistochemical staining was performed according to the manufacturer’s protocol of the SABC (goat-IgG)-POT kit and P-V6000 kit. Briefly, 4-μm sections were deparaffinized with xylene, rehydrated through a graded alcohol series, and rinsed in phosphate-buffered saline (PBS). Antigen retrieval was performed by pacing the slides in boiling citric acid buffer at pH 6.0 for 5 min. The sections were sequentially managed in 3 % H2O2 in methanol for 15 min at room temperature to quench endogenous peroxidase activity and then were incubated overnight at 4 °C with the primary antibodies of ING4 (1:400, goat polyclonal; abcam, USA) and Ki67, CD117, CD34, S100, Dog-1, desmin, vimentin (mouse monoclonal; Zhongshanjin-bio Co.). To the ING4 sections, biotin-labeled goat secondary antibody was added by drops, followed by further incubation with streptavidin–horseradish peroxidase. To the other sections, the biotin-labeled secondary antibodies only were added at room temperature for 30 min. Intervening PBS washing was necessary between each two steps. Immunoreaction was visualized with diluted DAB, and the sections were counterstained with hematoxylin and mounted.
Evaluation of immunohistochemical staining
The immunohistochemical staining for ING4 was mainly located in the nucleus and/or cytoplasm. Positive staining presented brown-yellow particles, and no staining was negative. A semiquantitative evaluation system was adopted to evaluate the staining effects. Five random high-power fields or 500 tumor cells were selected to count positively stained cells. The staining intensity was classified by the following criteria: 0 (no staining), 1 (light yellow/weak staining), 2 (yellow-brown/moderate staining), or 3 (brown/strong staining). The proportion of positively stained tumor cells was scored as follows: 0 (no positive tumor cells), 1 (positive tumor cells <10 %), 2 (positive tumor cells 10–50 %), 3 (positive tumor cells 51–75 %), or 4 (positive tumor cells >75 %). Staining index was calculated as the staining intensity score × the proportion score. A staining index score of six was used to distinguish low and high expression of ING4.
The immunohistochemical staining for Ki67 is mainly located in the nuclei of proliferative tumor cells. The percentage of positively stained nuclei was calculated by counting 10 randomly selected microscopic fields under high-power magnification. Finally, the samples were divided into two groups: a Ki67-negative group (<10 %) and a Ki67-positive group (≥10 %).
The immunohistochemical staining for CD117, CD34, Dog-1, SMA, S100, vimentin, and desmin was mainly localized in the cytoplasm, usually with diffuse distribution: these were used in the diagnosis of GISTs.
All the immunohistochemical staining sections was assessed and scored independently by two investigators. To ensure experimental accuracy, one of the investigators is a histopathology expert.
Statistical analysis
All data were analyzed by statistics software (SPSS 19.0 for Windows; SPSS). The chi-square and Mann–Whitney tests were used to compare the levels of ING4 expression in different risk GISTs and various clinicopathological parameters. The correlations between two independent variables were analyzed by calculating the Spearman’s correlation coefficients. Survival analysis was performed by the Kaplan–Meier method and log-rank test. The prognostic relevance was evaluated by Cox regression analysis. All measurement data were defined by mean ± SD. Paired comparison was demonstrated by t test. p < 0.05 was considered as significant.
Results
Diagnosis and clinicopathological features of GISTs
Forty-one patients with GISTs, 20 men and 21 women, were diagnosed by H&E stain and a panel of immunohistochemical staining for CD117, CD34, Dog-1, SMA, S-100, desmin, and vimentin (Fig. 1). The clinicopathological features of all patients with GISTs are listed in Table 2. The patients were followed up for 1 to 77 months, with median follow-up 53 months. Fifteen of 41 patients died during this period, a mortality rate of 36.6 %; tumor necrosis was found in 31.7 % of samples (13/41); mitotic index per 50 high-power fields was 17.63 ± 19.85; invasion rate was 34.1 % (14/41); recurrence and metastasis rate was 31.7 % (13/41); mean tumor size was 6.83 ± 5.34; and 22 of 41 GISTs (53.7 %) were high-risk tumors. The differences of lower- and high-risk groups are displayed in Table 3.
Hematoxylin and eosin (H&E) and immunohistochemical technique used to diagnose gastrointestinal stromal tumors (GISTs). a, b H&E stain. In our samples, immunostains for CD117 (c, d), Dog-1 (e, f), and vimentin (g, h) were all positive. Immunostain for CD34 was 78 % positive (i, j). Immunostains for S100 (k, l), smooth muscle actin (SMA) (m, n), and desmin (o, p) were all negative. a, c, e, g, i, k, m, o ×100; b, d, f, h, j, l, n, p ×400
Low expression of ING4 and high expression of Ki67 by IHC was associated with the high-risk group of GISTs
In the high-risk group, 100 % (22/22) samples showed decreased expression of ING4, and the expression level was 9.23 ± 7.66. In the lower-risk group, however, 89.5 % (17/19) samples displayed high ING4 expression, and the expression level was 78.95 ± 27.90. As shown in Tables 3, 4, and 5, and Fig. 2, the lower-risk group presented with a higher expression of ING4 compared with the high-risk group. There was a decreased tendency in the expression of ING4 from lower-risk to high-risk GISTs. However, an opposite tendency was observed in the expression of Ki67 (Tables 3, 5; Fig. 3). We further evaluated the correlation of ING4 and Ki67 (Table 6). In the low ING4 expression group, Ki67 expression was high (20/24), with an expression level of 17.63 ± 9.29. However, in the high ING4 expression group, the Ki67 expression was low (16/17), with an expression level of 4.00 ± 3.30. There was a strong negative correlation in the expression level of ING4 and Ki67 (χ2 = 23.892, p < 0.001).
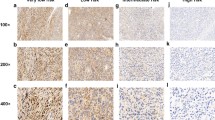
Expression level of ING4 by immunohistochemical technique in various risk GISTs: lower-risk group (a ×100; b ×400); high-risk group (c ×100; d ×400). ING4 showed significantly low expression in high-risk GISTs compared with lower-risk GISTs, with a statistically significant difference (p < 0.001). Expression of ING4 was mostly lost in high-risk GISTs, but the normal fiber and blood vessels around the tumor cells had high expression (e ×100; f ×400)
Low expression of ING4 was associated with the high-risk group of GISTs by univariate analysis
In an effort to predict GIST behavior, we performed a univariate analysis of ING4 expression. Several factors were related to clinical outcome including age, gender, tumor location, tumor size, cytotype, mitotic index, tumor necrosis, invasion, mortality, recurrence, and metastasis (Table 7). Low expression of ING4 expression was correlated with tumor location (small intestine, mesentery, omentum, and retroperitoneum) (p = 0.008), tumor size (≥5 cm; p < 0.001), mitotic index (≥5/50 HPF; p < 0.001), tumor necrosis (p = 0.021), invasion (p < 0.001), recurrence or metastasis (p = 0.021); and mortality (p < 0.001) of GISTs. Its expression was not correlated with age (p = 0.105), gender (p = 0.279), and cytotype (p = 0.066). Spearman correlation analysis was applied to further confirm the dependability between ING4 expression, Ki67 expression, and the clinicopathological characteristics. The results of this analysis are as follow: tumor location, 0.388 (p = 0.012); tumor size, −0.623 (p < 0.001); mitotic index, −0.852 (p < 0.001); tumor necrosis, −0.361 (p = 0.021); invasion, −0.606 (p < 0.001); recurrence and/or metastasis, −0.361 (p = 0.021); mortality, −0.639 (p < 0.001); and expression of Ki67, −0.763 (p < 0.001).
Low expression of ING4 was strongly associated with poor survival time of patients with GISTs
Kaplan–Meier analysis and log-rank test were used to evaluate the effects of ING4 expression on recurrence, metastasis, and mortality of GISTs. The log-rank test revealed a significant difference in survival time between the low and high ING4 expression groups (χ2 = 17.161, p < 0.001). High expression of ING4 was correlated with longer survival time of patients (p < 0.001), with a correlation coefficient of 0.601. In fact, the 5-year cumulative survival rate was 100 % in the high ING4 expression group but only 37.5 % in the low ING4 expression group (Fig. 4).
To determine whether ING4 could be a prognostic factor of GISTs, we did further multivariate survival analysis, using Cox regression in different risk groups (p = 0.004), invasion (p = 0.002), recurrence and/or metastasis (p = 0.014), and ING4 expression (p = 0.014) separately. Our findings indicated that ING4 could serve as a prognostic factor of GISTs.
Discussion
ING4, a member of the ING family, was first identified by Shiseki et al. [8]. In 2004, ING4 was defined as an important inhibitor of tumor growth [16]. It can induce tumor cell apoptosis by enhancing p53 transcription, which in turn can shorten the S phase of RKO colon cancer cells and enhance the G2/M-phase arrest of HepG2 liver cancer cells [23, 27, 28]. Moreover, it can inhibit the growth of glioma and associated angiogenesis by interacting with the P65 (RelA) subunit of NF-κB to inhibit its transcription and further downregulate the downstream genes of NF-κB, such as IL-8 and Cox-2 [16]. ING4 also can enhance the sensitivity of liver cancer cells to chemical agents that can damage DNA [28, 29], restore the contact inhibition among tumor cells [30], and inhibit hypoxia-inducible factor (HIF) activation by combining with HIF-α1, which specifically inhibits angiogenesis and tumor growth [29]. ING4 is involved in tumor cell apoptosis by activation of the mitochondrial-induced apoptotic pathway [23, 31] and regulation of Wnt-1/β-catenin signaling [18, 32]. Many studies showed that the expression of ING4 was obviously decreased or the ING4 gene was deleted in many human malignant tumors [23, 32]. Kim et al. [23] examined ING4 transcripts for mutations that might affect the gene product by cloning the coding sequence of ING4 transcripts with reverse transcriptase-polymerase chain reaction (RT-PCR) and sequencing at least two independent clones from each cell line. They found that mutations in ING4 transcripts were prevalent: seven of nine cancer cell lines contained mutant transcripts. Several cell lines contained a mutation that deletes parts of a nuclear localization signal (KGKK). Other cell lines contained single-nucleotide deletions that result in the C-terminal truncation of the ING4 proteins. Such truncations have a dominant-negative effect, which has the potential to create a null state in even those cells that are heterozygous for the wild-type ING4 gene. In addition, their analysis of comparative genomic hybridization (CGH) data revealed that 10–20 % of primary breast tumors have chromosome deletions in 12p13, and the deletions appear to affect only one copy of the gene; no genomic mutations were, however, found in the remaining allele of ING4. Reduction in the ING4 transcript level caused by a single copy deletion may contribute to tumorigenesis, or ING4 transcripts from the remaining allele may contain inactivating alterations such as the KGKK deletion mutation. Gunduz et al. [17] demonstrated a frequently deleted region that is more telomeric to polymorphic marker D12S89 in head and neck squamous cell carcinoma (HNSCC). The highest loss of heterozygosity (LOH) (56 %) was found at the marker D12S825, which is very near ING4. Moreover, flanking markers D12S77 and D12S372 also showed a very high LOH ratio. This frequently deleted region was bounded by the markers D12S372 and D12S77, and the central marker D12S825 is just about 700 bp away from ING4. This region is heavily populated with many genes. Because ING4 is localized in this region and other members of ING4 were previously shown to be TSGs (tumor suppressor genes), they found it to be a strongly targeted candidate in their samples. In addition, mutation analysis was performed at mRNA level using cDNAs prepared from tumor samples, and therefore mutant mRNAs may not be stable and could be easily degraded. Furthermore, decreased expression of ING4 was detected in 76 % of tumor samples as compared with their normal counterparts with quantitative real-time PCR; most of those samples with low ING4 expression displayed extremely low mRNA expression. Last, high expression of ING4 was correlated with well-differentiated tumors whereas low expression of ING4 was seen with poorly differentiated tumors [18]. Cai et al. [32] used the immunohistochemical technique with specific antibodies against ING4 to examine the level of ING4 in melanomas and benign melanocytic nevi, finding that the expression of ING4 was significantly reduced in the melanoma cells compared with benign melanocytic nevi and normal skin tissues. However, in the literature, there have been no publications regarding ING4 expression status in GISTs. In our study, the results were similar to the previous studies in other tumor types: the high-risk GISTs (100 %, 22/22) showed markedly decreased ING4 protein with an expression level of 9.23 ± 7.66, and the lower-risk GISTs (89.5 %, 17/19) displayed high ING4 protein with an expression level of 78.95 ± 27.90. There was a significant difference in ING4 expression level between lower-risk and high-risk GISTs (p < 0.001). The expression level of ING4 was positively correlated with the tumor cell differentiation of GISTs. The reduced expression or loss of function through tumor suppressor gene alterations is an important mechanism in the progression of malignant tumors. Our results suggest ING4 downregulation to be involved in the tumor progression of GISTs. However, the molecular mechanisms of ING4 downregulation in GISTs are not clear. Possibilities of ING4 gene alterations or epigenetic regulations include mutations, deletions, and LOH. Therefore, further mechanistic studies are required to uncover the causes of ING4 protein downregulation in GISTs.
To further confirm the associations of ING4 expression and the tumor progression of GISTs, we examined Ki67 expression in these samples. The expression level of Ki67 was 4.42 ± 3.75 in lower-risk GISTs and 18.50 ± 9.09 in high-risk GISTs. A significant difference was found between lower-risk and high-risk GISTs (p < 0.001). Spearman correlation analysis, applied to further confirm the interdependability between the expression of ING4 and Ki67 with the correlation value of −0.763 (p < 0.001), indicated the expression of ING4 was negatively correlated with the expression of Ki67. Ki67, an important proliferation-associated protein, has already been demonstrated to show high expression in many malignant tumors. Also, it is correlated with tumor progression, metastasis, and prognosis. Our results indicate that both ING4 and Ki67 are correlated with GIST tumor progression, metastasis, and prognosis.
The major cause of death in patients with malignancies is invasion or metastasis. NF-κB has been shown to facilitate angiogenesis and metastasis of malignant tumors. Some studies indicated that ING4 physically interacts with the p65 (RelA) subunit of nuclear factor NF-κB, and ING4 regulates tumor angiogenesis through transcriptional repression of NF-κB-responsive genes [16] and, therefore, suppresses angiogenesis and metastasis. ING4 could also suppress tumor cell movement by interacting with liprin α1 protein, which is a cytoplasmic protein necessary for focal adhesion formation and axon guidance [21], and inhibit tumor cell infiltration and metastasis [19, 22]. The effect of ING4 protein expression on invasion and metastasis of GISTs has not been clearly addressed yet. We did a further analysis of the correlation between ING4 expression and survival factors of patients with GISTs. We found that ING4 scores were significantly higher in the non-invasion group than in the invasion group (median values, 9.93 and 57.93; p < 0.001) and in the no-recurrence-or-metastasis group than in the recurrence and/or metastasis group (median values, 18.15 and 52.39; p < 0.001). These results indicated that the expression of ING4 had significant correlation with the survival factors of GISTs (such as invasion, recurrence, and metastasis). We speculate that ING4 might interact with NF-κB and/or liprin α1 to inhibit tumor cell invasion and recurrence and metastasis of GISTs. This point needs further investigation.
The examination of CD117/c-KIT by immunohistochemistry has become a prognostic criterion for GISTs now, but it is not an ideal prognostic indicator. DeMatteo et al. [33] reported that only tumor size was used as a predictive indicator of GIST prognosis in their research. Other prognostic indicators for GISTs reported in the literature include the number of mitotic figures per 50 HPF in addition to tumor size and Ki67 proliferation index [4, 34]. Similar results were found in our research, that both tumor size and mitotic figures per 50 HPF related to the prognosis of GISTs. Moreover, the multivariate survival analysis revealed the 5-year survival in the high expression group of ING4 is 100 % and in the low expression group is 38 %, with a significant difference in the two groups (p < 0.001). The results suggested the expression level of ING4 was negatively correlated with prognosis of GISTs. Low expression of ING4 correlated with poor prognosis of GISTs. In addition, a recent study has identified CD26 associated with the increased risk of postoperative recurrence warrants diagnostic application, but only CD26 is associated with the outcome of the gastric GISTs [35]. In our results, tumor location has statistical significance in different ING4 expression groups. Many previous studies concluded that small intestinal GISTs have a worse outcome than gastric GISTs. Therefore, we did further analysis to determine whether the tumor location has an effect on the outcome of the patients in our study. Our results indicate that tumor location does not have an effect on the survival of the patients in our research (Cox regression; p value = 0.070). It is implied that ING4 might be as an indicator of GIST prognosis no matter where the tumor arose.
In conclusion, our study indicates that ING4 most likely plays an important role in inhibiting the tumor progression of GISTs. ING4 is, therefore, a good candidate both as a clinically relevant indicator of disease progression and as a prognostic biomarker for survival of patients with GISTs.
References
Miettinen M, Lasota J. Gastrointestinal stromal tumors—definition, clinical, histological, immunohistochemical, and molecular genetic features and differential diagnosis. Virchows Arch. 2001;438:1–12.
Blanke CD, Corless CL. State-of-the art therapy for gastrointestinal stromal tumors. Cancer Invest. 2005;23:274–80.
Hou YY, Tan YS, Sun MH, Wei YK, Xu JF, Lu SH, et al. C-kit gene mutation in human gastrointestinal stromal tumors. World J Gastroenterol. 2004;10:1310–4.
Miettinen M, Sarlomo-Rikala M, Lasota J. Gastrointestinal stromal tumors: recent advances in understanding of their biology. Hum Pathol. 1999;30:1213–20.
Miettinen M, Sarlomo-Rikala M, Sobin LH, Lasota J. Gastrointestinal stromal tumors and leiomyosarcomas in the colon: a clinicopathologic, immunohistochemical, and molecular genetic study of 44 cases. Am J Surg Pathol. 2000;24:1339–52.
Miettinen M, Majidi M, Lasota J. Pathology and diagnostic criteria of gastrointestinal stromal tumors (GISTs): a review. Eur J Cancer. 2002;38(suppl 5):S39–51.
Stamatakos M, Douzinas E, Stefanaki C, Safioleas P, Polyzou E, Levidou G, Safioleas M. Gastrointestinal stromal tumor. World J Surg Oncol. 2009;7:61.
Shiseki M, Nagashima M, Pedeux RM, Kitahama-Shiseki M, Miura K, Okamura S, et al. p29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity. Cancer Res. 2003;63:2372–8.
Garkavtsev I, Kazarov A, Gudkov A, Riabowol K. Suppression of the novel growth inhibitor P33ING1 promotes neoplastic transformation. Nat Genet. 1996;14:415–20.
Chen L, Matsubara N, Yoshino T, Nagasaka T, Hoshizima N, Shirakawa Y, et al. Genetic alterations of candidate tumor suppressor ING1 in human esophageal squamous cell cancer. Cancer Res. 2001;61:4345–9.
Garkavtsev I, Riabowol K. Extension of the replicative life span of human diploid fibroblasts by inhibition of the p33ING1 candidate tumor suppressor. Mol Cell Biol. 1997;17:2014–9.
Nagashima M, Shiseki M, Pedeux RM, Okamura S, Kitahama-Shiseki M, Miura K, et al. A novel PHD-finger motif protein, p47ING3, modulates p53-mediated transcription, cell cycle control, and apoptosis. Oncogene. 2003;22:343–50.
Wagner MJ, Gogela-Spehar M, Skirrow RC, Johnston RN, Riabowol K, Helbing CC. Expression of novel ING variants is regulated by thyroid hormone in the Xenopus laevis tadpole. J Biol Chem. 2001;276:47013–20.
Gunduz M, Ouchida M, Fukushima K, Ito S, Jitsumori Y, Nakashima T, et al. Allelic loss and reduced expression of the ING3, a candidate tumor suppressor gene at 7q31, in human head and neck cancers. Oncogene. 2002;21:4462–70.
Hu RM, Han ZG, Song HD, Peng YD, Huang QH, et al. Gene expression profiling in the human hypothalamus-pituitary-adrenal axis and full-length cDNA cloning. Proc Natl Acad Sci USA. 2000;97:9543–8.
Garkavtsev I, Kozin SV, Chernova O, Xu L, Winkler F, Brown E, et al. The candidate tumor suppressor protein ING4 regulates brain tumor growth and angiogenesis. Nature (Lond). 2004;428:328–32.
Gunduz M, Nagatsuka H, Demircan K, Gunduz E, Cengiz B, Ouchida M, et al. Frequent deletion and down-regulation of ING4, a candidate tumor suppressor gene at 12p13, in head and neck squamous cell carcinomas. Gene (Amst). 2005;356:109–17.
Li XM, Cai LM, Liang MH, Wang YD, Yang J, Zhao YL. ING4 induces cell growth inhibition i human lung adenocarcinoma A549 cells by means of Wnt-1/β-catenin signaling pathway. Anat Rec. 2008;291:593–600.
Li J, Martinka M, Li G. Role of ING4 in human melanoma cell migration, invasion and patient survival. Carcinogenesis (Oxf). 2008;29:1373–9.
Fang F, Luo LB, Tao YM, Wu F, Yang LY. Decreased expression of inhibitor of growth 4 correlated with poor prognosis of hepatocellular carcinoma. Cancer Epidemiol Biomark Prev. 2009;18:409–16.
Unoki M, Shen JC, Zheng ZM, Harris CC. Novel splice variants of ING4 and their possible roles in the regulation of cell growth and motility. J Biol Chem. 2006;281:34677–86.
Shen JC, Unoki M, Ythier D, Duperray A, Varticovski L, Kumamoto K, et al. Inhibitor of growth 4 suppresses cell spreading and cell migration by interacting with a novel binding partner, liprin alpha1. Cancer Res. 2007;67:2552–8.
Kim S, Chin K, Gray JW, Bishop M. A screen for genes that suppress loss of contact inhibition: identification of ING4 as a candidate tumor suppressor gene in human cancer. Proc Natl Acad Sci USA. 2004;101:16251–6.
Ozer A, Wu LC, Bruick RK. The candidate tumor suppressor ING4 represses activation of the hypoxia inducible factor (HIF). Proc Natl Acad Sci USA. 2005;102:7481–6.
Fletcher C, Berman J, Corless C, Gorstein F, Lasota J, Longley BJ, et al. Diagnosis of gastrointestinal stromal tumors: a consensus approach. Hum Pathol. 2002;33:459–65.
Fletcher C, Berman J, Corless C, Gorstein F, Lasota J, Longley BJ, et al. Diagnosis of gastrointestinal stromal tumors: a consensus approach. Int J Surg Pathol. 2002;10:81–9.
Zhang X, Wang KS, Wang ZQ, Xu LS, Wang QW, Chen F, et al. Nuclear localization signal of ING4 plays a key role in its binding to p53. Biochem Biophys Res Commun. 2005;331:1032–8.
Zhang X, Xu LS, Wang ZQ, Wang KS, Li N, Cheng ZH, et al. ING4 induces G2/M cell cycle arrest and enhances the chemosensitivity to DNA-damage agents in HepG2 cells. FEBS Lett. 2004;570:7–12.
Colla S, Tagliaferri S, Morandi F, Lunghi P, Donofrio G, Martorana D, et al. The new tumor-suppressor gene inhibitor of growth family member 4 (ING4) regulates the production of proangiogenic molecules by myeloma cells and suppresses hypoxia-inducible factor-1 alpha (HIF-1alpha) activity: involvement in myeloma-induced angiogenesis. Blood. 2007;110:4464–75.
Li XM, Zhang QY, Cai LM, Wang YH, Wang Q, Huang XY, et al. Inhibitor of growth 4 induces apoptosis in human lung adenocarcinoma cell line A549 via Bcl-2 family proteins and mitochondria apoptosis pathway. J Cancer Res Clin Oncol. 2008;135:829–35.
Li XM, Cai LM, Chen H, Zhang QY, Zhang SJ, Wang YH, et al. Inhibitor of growth 4 induces growth suppression and apoptosis in glioma U87MG. Pathobiology. 2009;76:181–92.
Cai LM, Li XM, Zheng SY, Wang YH, Wang YD, Li HY, et al. Inhibitor of growth 4 is involved in melanomagenesis and induces growth suppression and apoptosis in melanoma cell line M14. Melanoma Res. 2009;19:1–7.
DeMatteo RP, Lewis JJ, Leung D, Mudan SS, Woodruff JM, Brennan MF. Two hundred gastrointestinal stromal tumors: recurrence patterns and prognostic factors for survival. Ann Surg. 2000;231:51–8.
Sandberg AA, Bridge JA. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors. Synovial sarcoma. Cancer Genet Cytogenet. 2002;133:1–23.
Yamaguchi U, Nakayama R, Honda K, Ichikawa H, Hasegawa T, Shitashige M, et al. Distinct gene expression-defined classes of gastrointestinal tumor. J Clin Oncol. 2008;26:4100–8.
Acknowledgments
This study was from Project No. D201201, supported by the Natural Science Foundation of Heilongjiang Province of China. It was also supported by the postdoctoral scientific research start-up fund of Heilongjiang Province of China (No. LBH-Q11070).
Author information
Authors and Affiliations
Corresponding authors
Additional information
A. Nanding, L. Tang, L. Cai, and H. Chen contributed equally to this work.
Rights and permissions
About this article
Cite this article
Nanding, A., Tang, L., Cai, L. et al. Low ING4 protein expression detected by paraffin-section immunohistochemistry is associated with poor prognosis in untreated patients with gastrointestinal stromal tumors. Gastric Cancer 17, 87–96 (2014). https://doi.org/10.1007/s10120-013-0248-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10120-013-0248-8