Abstract
Climate change is projected to influence the genetic resources of plant species. Recent research has examined genetic diversity patterns under current climate conditions, with little attention to the future genetic consequences for species. In this study, we combined ecological niche modeling and population genetic approaches to project future changes in genetic diversity using plastid and nuclear DNA and reconstructed distribution patterns of three circumboreal plants (Chamaedaphne calyculata, Linnaea borealis ssp. borealis, and Pedicularis sceptrum-carolinum ssp. sceptrum-carolinum) in the last glacial maximum. We found that circumboreal plants could potentially lose their geographic ranges in the future (2070; 35–52% in RCP 4.5 (representative concentration pathways), 37–53% in RCP 6.0, and 56–69% in RCP 8.5), only slightly compensated by a predicted range gain of 18–33% (across the three RCPs). It is expected that future genetic diversity level could remain similar or lower than the present level. On the other hand, the homogeneity of the genetic background—a lack of admixture and domination of one gene pool in most populations of C. calyculata and L. borealis ssp. borealis—was predicted to become more pronounced in the future. Combining the paleoecological niche modeling and genetic data revealed, more precisely, the climate refugia for circumboreal plants in the Alps, central Asia, Beringia, and southern North America and the macrorefugia more restricted to the northern part of Eurasia and North America, reaching the arctic zone.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Life-history traits, population dynamics, and natural or human impacts (e.g., isolation, fragmentation, and climate changes) have been widely discussed as major determinants of intraspecific genetic diversity (e.g., Loveless and Hamrick 1984; Hamrick and Godt 1989; Barrett and Husband 1990; Young et al. 1996; Gitzendanner and Soltis 2000; Nybom 2004; Jump and Peñuelas 2006; Parmesan 2006; Bizoux and Mahy 2007). In recent years, climate has been more frequently considered as a factor shaping genetic resources within species, through species distribution shifts and increasing range fragmentation. Although surveys of genetic diversity under current climate conditions have a rich history in the literature, potential future impacts on the genetic diversity of species are rarely examined (Alsos et al. 2009; Thomassen et al. 2010; Beatty and Provan 2011; Collevatti et al. 2011; Alsos et al. 2012; Jay et al. 2012; Eidesen et al. 2013; Temunovic et al. 2013; Steane et al. 2014). Northern plant species could be more severely threatened than other species by recent and future climate change (Thuiller et al. 2005; Parmesan 2006). Only Alsos et al. (2012) have exhaustively described future genetic and geographic diversity patterns of northern plants; they pointed out that a higher loss of genetic diversity is to be expected within this plant group than for temperate species (Morin and Viner et Chuine 2008; Normand et al. 2009). Among the northern plants, arctic-alpine/alpine species are more sensitive than circumboreal species to potential loss of climatically suitable areas, because they are adapted to colder climates than the latter.
Among the factors that limit species distributions, undoubtedly the last glacial maximum (LGM, ca. 21 kya BP) was the major force shaping plant geographic ranges in the Northern Hemisphere. Considered separately, species distribution models and paleovegetation models have often yielded different putative distributions of northern organisms during the LGM. This has led to different conclusions about the geographic distribution of northern plants (Bhagwat and Wilis 2008; Allen et al. 2010; Beatty and Provan 2010; Svenning et al. 2011). Northern plants (boreal trees and shrubs) may have had wider geographic ranges during the LGM across Southern and Central Europe as well as in Central Asia. In North America, they may have been restricted to the narrow edge of the ice sheet and to fragments in different non-glaciated parts of continents (non-glaciated regions of Alaska and Yukon, southwestern and/or southeastern parts of North America). Different scenarios describing the phylogeographic history of northern plants during the late glacial and postglacial period indicate that even then, these species had geographic distributions similar to those of the present day (Tremblay and Schoen 1999; Abbott and Comes 2004; Jørgensen et al. 2006; Eidesen et al. 2007; Schönswetter et al. 2007; Ehrich et al. 2008; Keller et al. 2010). An understanding of how species’ genetic diversity was preserved during past climate change may give insights into the impact of such changes on present and future genetic diversity patterns. Recently, Gavin et al. (2014) combined studies of paleoecology, phylogeography, and species distribution models (SDMs) to identify species’ colonization routes and refugia during the LGM. This multi-faceted approach seems promising as an approach to understand species distributions in past and future.
Among northern plants, the genetic consequences of climate change may differ because their populations and refugial areas in Eurasia and North America occurred in different parts of the continents and genetic diversity was distributed in different ways. Alsos et al. (2012) estimated that in a third of 27 northern plants, more than 50% of their genetic diversity was expected to be lost under at least one emission scenario (e.g., A2 HadCM3) in 2080.
In this study, we integrated geospatial and climate information with data on genetic diversity patterns in order to more comprehensively examine the history of three circumboreal taxa: Chamaedaphne calyculata (Ericaceae), Linnaea borealis ssp. borealis (L. b ssp. borealis, Caprifoliaceae), and Pedicularis sceptrum-carolinum ssp. sceptrum-carolinum (P. s-c ssp. sceptrum-carolinum, Orobanchaceae). These taxa have partially overlapping and fragmented ranges in Eurasia as well as in North America (Meusel et al. 1978; Hultén and Fries 1986; Fig. 1a–c). Previous studies of these taxa did not consider the effects of climate change on their past and future geographic distributions and their future genetic resources, but focused on current phylogeographic and genetic diversity patterns in their Eurasian and North American geographic ranges (Wróblewska 2012, 2013a, b). Surveys of two plants (C. calyculata and L. b ssp. borealis) using cpDNA and ITS markers showed different phylogeographic patterns but surprisingly low Nei’s gene diversity in each of them (AFLP markers). A clear phylogeographic split between Eurasia/NW North America and NE North America was shown in the C. calyculata distribution, coinciding with Sphagnum-dominated peatlands in North America (Wróblewska 2012), while L. b ssp. borealis was characterized by a shallow and weak phylogeographic structure in its Eurasian range (Wróblewska 2013b). The genetic diversity of P. s-c ssp. sceptrum-carolinum was previously investigated only on a small range scale in western peripheral populations in Europe (Wróblewska 2013a). The taxa also differ in biology (growth form, type of reproduction, breeding system, and dispersal adaptation) and differ in population size within the geographic ranges (Wróblewska 2012, 2013a, b).
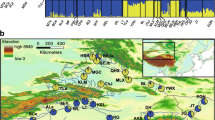
Ecological niche models (ENMs) for three circumboreal taxa. a Chamaedaphne calyculata. b Linnaea borealis ssp. borealis. c Pedicularis sceptrum-carolinum ssp. sceptrum-carolinum, distributed in Eurasia and North America for the last glacial maximum (LGM; ca. 21 kya BP; projected with CCSM4, MPI-ESM-P, and MIROC-ESM models), the present, and future (2070, projected with RCP 4.5, 6.0, and 8.5 scenarios). Color scale refers to logistic probability (0–1) of taxon occurrence, with color darkening with increasing climate suitability
One promising geospatial technique for use in evolutionary and ecological studies involves ecological niche models (ENMs; Svenning et al. 2011). These models indicate potential rather than realized distributions, but the difference between recent and potential future distributions can be a source of valuable information on possible range shifts (Guisan and Thuiller 2005), the risk status of current ranges, and the dispersal rates required to reach suitable new habitats (Jump et al. 2009). Based on ENM approaches, Jay et al. (2012) implemented ancestry distribution models to forecast changes in intraspecific genetic diversity in response to climate change, using correlations between environmental variables and individual ancestry. Ancestry distribution models can help predict the future genetic resources of species, which is important for assessing their evolutionary potential and for management. The circumboreal plants in the present study are considered to be under various levels of local extirpation due to the effects of climate change, and we expect their genetic resources to differ markedly in the future. We implemented ENM and population genetic approaches (i) to determine the respective climate envelopes for the past (ca. 21 kya BP), current, and potentially future (2070) geographic ranges within Eurasia and North America; (ii) to estimate changes between current and future population genetic structure in each taxon, that is, the current and future rates of gene migration correlated with environmental variables and changes in genetic diversity expected as a consequence of range loss; and to predict (iii) the climate refugia—locations where taxa survive periods of regionally adverse climate, areas where plants could survive during the LGM (ca. 21 kya BP, Ashcroft 2010; Gavin et al. 2014)—and the locations of their macrorefugia, that is, the overlap of current and future areas of suitable climate (Temunovic et al. 2013).
Materials and methods
Predictive ecological niche models for the last glacial maximum, present and future
For model training, we used available occurrence records of C. calyculata (N = 367) from Eurasia and North America, and of L. b ssp. borealis (N = 552) and P. s-c ssp. sceptrum-carolinum from Eurasia (N = 255, Supplementary information Fig. S1a–c). Combined with data extracted from the Global Biodiversity Information Facility (GBIF, http://www.gbif.org) as well as other databases (http://www.plantarium.ru and http://www.uaf.edu/museum/collections/herb/), literature, personal communications from collectors of samples, and our own sampling. In case of P. s-c ssp. sceptrum-carolinum, the occurrence records obtained were highly clustered in Scandinavia, which could overestimate the importance of the local conditions in the course of model training. Therefore, the occurrence records from this part of the range were reduced to one record per 2.5° of latitude and longitude. Such reduction aligned the spatial distribution of records used for model training (Supplementary information Fig. S1c). We used MAXENT v. 3.3.3e (Phillips et al. 2006) software to develop ENMs from 19 BIOCLIM variables related mostly to temperature and precipitation (http://www.worldclim.org/). First, we selected variables containing highly correlated pairs (Spearman rank correlation > 0.7; Giovanelli et al. 2010), since these can lead to overfitting of the model. After removing those variables that exhibited a strong correlation, five of them for C. calyculata and L. b ssp. borealis (BIO2 = mean diurnal range, BIO5 = max temperature of warmest month, BIO7 = temperature annual range, BIO8 = mean temperature of wettest quarter, BIO12 = annual precipitation) and seven of them for P. s-c ssp. sceptrum-carolinum (BIO1 = annual mean temperature, BIO3 = isothermality, BIO6 = min temperature of coldest month, BIO8 = mean temperature of wettest quarter, BIO9 = mean temperature of driest quarter, BIO11 = mean temperature of coldest quarter, BIO12 = annual precipitation) were used for modeling the current distribution and then reconstructing the paleodistribution during the LGM (ca. 21 kya BP) with CCSM4, MPI-ESM-P, and MIROC-ESM with spatial resolution between 2.5 min (of a longitude/latitude degree). ENMs were also projected to the future under climate change scenarios for 2070s. The climate change scenarios used for the projections followed three of the representative concentration pathways (RCPs) reported in the Coupled Model Intercomparison Project Phase 5 (CMIP5) data: RCP 8.5 (~ 8.5 W m−2 stabilized after 2100, slightly more severe than SRES A2), which corresponds to a “non-climate policy” scenario translating into high severity climate change impacts, a moderate mitigation scenario RCP 6.0 (stabilization at 6 W m−2 by 2100, similar to SRES A1B) requiring a climate-policy intervention, and a stronger mitigation scenario RCP 4.5 (~ 4.5 W m−2 stabilized after 2100, similar to the lowest-emission scenarios, B1) assuming the imposition of a series of emission mitigation policies (Masui et al. 2011; Riahi 2011; Thomson et al. 2011). Six CMIP5 models under three RCPs (8.5, 6.0, and 4.5) for the 2070s were used to avoid the bias related to the choice of a particular global climate model (GCM): CCSM4, HadGEM2-ES, IPSL-CM5A-LR, MIROC-ESM, MIROC5, and NorESM1-M. These models are among the most used models currently available for simulating the global climate response to increasing greenhouse gas concentration (Watanabe et al. 2011; Dufresne et al. 2013; Betts et al. 2015). Projections were summarized using the median of the predicted probabilities of occurrence across the six GCMs. Moreover, climatically suitable areas for studied populations were averaged for six GCMs used under three different RCP scenarios in order to show variance between models.
In ArcGIS v. 10.0 (ESRI Inc., Redlands, CA), we classified the probability of niche occurrence data in 10 equivalent classes of 0.1. In order to validate the model, the occurrence points were divided into two in two parts—75% of points were used to train the model and other 25% to test it. Models were evaluated upon the area under curve (AUC) value of receiver operator characteristics (ROC) for testing data. This is a common procedure, although with limitations (Lobo et al. 2008); therefore, values of test omission rates for selected threshold were also considered to judge model. There are several methods for selecting thresholds, and their possible impacts on the predicted distributions have been explored and discussed in the literature (Liu et al. 2005; Nenzén and Araújo 2011). However, there is currently no consensus on the most suitable and stable method for applying thresholds to species range projections (Nenzén and Araújo 2011). The logistic output in MaxEnt estimates the probability of presence, assuming that the sampling design is such that typical presence localities have a presence probability of ca. 0.5, but this value is fairly arbitrary (Liu et al. 2005; Phillips et al. 2006). Therefore, we selected those catchments corresponding to MaxEnt values indicating maximum training sensitivity plus specificity, which is considered a more robust approach (Liu et al. 2005).
We also presented the overall (past, present, and future) models as maps of overlap between the current and predicted geographic ranges (2070) to visualize putative macrorefugia under climate change (Temunovic et al. 2013). The reduction of climatically suitable areas across the future geographic range was calculated as the frequency of current pixels to be lost in 2070.
Sample collection and genetic diversity under scenarios of current and future climate change
Three circumboreal plant taxa were collected from their geographic range, making a total of 769 plant samples from 68 populations. The sampling strategy has been published in Wróblewska (2012, 2013a, b). The details of sequencing and AFLP laboratory procedures have been published in Wróblewska (Wróblewska 2012, 2013a, b; Supplementary information Table S1).
For the phylogenetic analysis, sequences were imported into BEAST 2.0 software (Drummond et al. 2012). We used the BEAUTI program to unlink the substitution models of the data partitions and to implement the models of sequence evolution identified as optimal by JMODELTEST 0.1.1 (Posada 2008). The fit of the nucleotide substitution models was assessed with the Akaike information criterion (AIC). Simulations were run using Markov Chain Monte Carlo (MCMC) for 10,000,000 generations, with a storage of 10,000 in BEAUTI. Finally, we discarded the trees as burn-in and summarized the trees using TREEANNOTATOR 1.7.2. The tree was visualized with FIGTREE 1.3.1 (Rambaut 2010).
We standardized the minimum sample size using the rarefaction as the rarefy function of R package vegan (Mitka et al. 2015; Oksanen et al. 2015) and then calculated mean intra-population gene diversity (H, Nei 1987) using function diversity and frequency-down-weighted marker values (DW) using function rarity index as described by Schönswetter and Tribsch (2005) with AFLPdat software (Ehrich 2006) in each population of the three taxa. We assumed that only populations found in regions above a certain level of suitability (above the value of maximum training sensitivity plus specificity threshold) will persist. It is possible to recalculate the overall genetic diversity parameters (H and DW) within each plant across the geographic range, which generates the gene pool of individuals for the future generations. Spearman rank correlations were used to estimate the degree of association of gene diversity (H) and frequency-down-weighted marker values (DW) with longitude and latitude and bioclimatic variables for modeling the current distribution for each taxon. We also estimated the expected loss of genetic diversity within each investigated taxon as the consequences of range loss based on the assumptions and method proposed by Alsos et al. (2012). The study area was divided on an arbitrary grid of 500 km × 500 km cells using ARCMAP 9.2, and only grid cells with the sampling localities were considered. In this method, the loss of genetic diversity is considered as the loss of AFLP markers occurring from randomly removing an increasing number of grid cells. The results were then plotted showing minimum, maximum, mean, and median number of markers lost for an increasing number of grid cells removed. It was shown to estimate for differences in sampling intensity in different areas.
Genetic differentiation among the sampled populations within each species (F ST) was determined by analyses of molecular variance (AMOVA), and levels of significance for populations were determined using a permutation test (1000 permutations) to obtain solid test statistics with ARLEQUIN 3.11 (Excoffier et al. 2005).
For each plant taxon, to infer population structure, we used the spatial Bayesian clustering algorithm described by Jay (2011) and implemented in POPS (http://membres-timc.imag.fr/Olivier.Francois/pops.html). This Bayesian algorithm incorporates genetic and geographic data and infers an admixture coefficient based on the correlation between environmental variables and individual ancestry (Jay 2011). POPS also provides a facility to estimate changes in population genetic structure under a scenario assuming future climate changes that can provide indirect estimates of future rates of gene migration (Jay et al. 2012).
We studied plants in which both sexual and vegetative reproduction could occur. Previous studies of these three taxa revealed that multidirectional gene flow was possible and not limited within the geographic range (Wróblewska 2012, 2013a, b); therefore, we consider admixture models (Falush et al. 2007). Ten independent replicates were run for all possible values of the maximum number of clusters (K max ) in each plant. We ran POPS using the spatial coordinates of each individual as covariates. The forecasts from regression models were obtained from bioclimatic variables for the current climate, considering the same variables selected from ENM, for three RCPs. For the simulated data, the POPS runs used 200,000 sweeps with an initial burn-in of 100,000 sweeps. We identified the optimal genetic structure when the deviance information criterion (DIC) curve began to plateau. Then, we applied the same genetic and geographic data, while the same bioclimatic variables were generated for 2070 to forecast the population genetic structure in the future for each taxon. In C. calyculata, we identified two phylogeographic groups within the geographic range (coinciding with Sphagnum-dominated peatlands); so, we performed a spatial Bayesian clustering algorithm (Jay 2011) in these groups separately.
Results
Ecological niche model
In our data, the AUC values indicate very high model fit for each studied species. The variance between the models represented about 30–50% of the mean climatically suitable areas and was comparable between RCP scenarios, but lower in P. s-c ssp. sceptrum-carolinum than in other two studied species (Supplementary information Table S2). For C. calyculata, L. b ssp. Borealis, and P. s-c ssp. sceptrum-carolinum, the respective AUC training values pertaining to the ROC curve produced by MAXENT were 0.880, 0.867, and 0.937, and the testing values were 0.906, 0.892, and 0.966. Test omission rates reached moderate level (0.231) for C. calyculata and fairly accurate (0.135) for L. b ssp. borealis and (0.026) P. s-c ssp. sceptrum-carolinum, respectively. The truncation threshold was 0.342 for C. calyculata, 0.395 for L. b ssp. borealis, and 0.178 for P. s-c ssp. sceptrum-carolinum; regions with suitability values lower than those given above for each taxon were unlikely to maintain the taxa in any scenario. We generated an ecological niche model (ENM) of the current range (Fig. 1a–c), which described relatively well the present distribution of C. calyculata, L. b ssp. borealis, and P. s-c ssp. sceptrum-carolinum. For L. b ssp. borealis, the current modeled distribution also reached northern North America, where only the distribution ranges of other L. b ssp. borealis subspecies have been reported (ssp. americana (Forbes) Hultén ex R.T. and ssp. longifolia (Torr.) Hultén). The paleo-ENMs were more variable between circumboreal species. A wider range of suitable areas for persistence during the LGM was identified in C. calyculata in southern areas of Europe, Southeast and East Asia, and southern North America, as well as central Alaska and part of Yukon Territory (Fig. 1a); in L. b ssp. borealis—such areas occurred at 48–69° N and in central Alaska (1b). In P. s-c ssp. sceptrum-carolinum, the paleo-ENM showed that Southeast Asia and the more southward parts of Europe became probable suitable climate refugia during the LGM (Fig. 1c).
Future ENMs predicted that climate change will result in a clear northward shift in the distribution of the studied circumboreal plants, with suitable conditions restricted to northern parts of Eurasia and/or North America. Future suitable areas also spread to the Tibetan Plateau (Fig. 1a–c). The predicted future range of P. s-c ssp. sceptrum-carolinum will be more restricted than for the other two, with a narrow northern zone in Europe in both simulations. With the threshold indicating sensitivity-specificity sum maximization, the shares of geographic ranges that may be lost in the future were quite similar: for L. b ssp. borealis and P. s-c ssp. sceptrum-carolinum (52%) and lower in C. calyculata (35%) in RCP 4.5, from 37 to 53% in RCP 6.0 and more severe 56–69% in RCP 8.5 (Table 1). The percentage of range gain for 2070 was similar for C. calyculata (19–29%) and for L. b ssp. borealis (ca. 18–29%) and slightly higher for P. s-c ssp. sceptrum-carolinum (21–33%) in all RCP scenarios. Strong range turnover is predicted for all. The current range will overlap with the future model from 32% (RCP 8.5) to 54% (RCP 6.0) in C. calyculata, by 27% (RCP 8.5) and 38% (RCP 6.0) in L. b ssp. borealis, and by 25% (RCP 8.5) to 48% (RCP 6.0) in P. s-c ssp. sceptrum-carolinum (Table 1). On the maps, the overlaps of the current and predicted ranges indicate putative refugia from climate change (macrorefugia) in northernmost Europe for all studied plants (Fig. 1a–c). In Asia, the overlaps for each plant are mainly in the narrow northern zone of the current and future geographic ranges, and in North America for C. calyculata.
Genetic diversity resources under current and future scenarios
DNA sequencing
We identified seven cpDNA haplotypes in C. calyculata (Fig. 2a; Table 1, Supplementary information Table S3). Two cpDNA haplotypes were detected exclusively in the Eurasian/NW North American group. Five cpDNA haplotypes (C-G) were found only along NE North America, and three of them were unique (Fig. 2a). We detected 35 ITS haplotypes (Fig. 2a, Table 1, Supplementary information Table S2). The Eurasian/NW North American group consisted of 17 haplotypes belonging to 19 populations; 18 haplotypes were found exclusively in ten populations from NE North America. The rooted strict Bayesian phylogenetic reconstruction based on cpDNA markers revealed one well-supported Eurasian/NW North American group (99%, Fig. 2a). For ITS sequences, the tree supported only the NE North American group (94%, Fig. 2a).
Distribution of cpDNA and ITS haplotypes at the present time and their phylogenetic relationships (inferred from BEAST) are shown in the inset within a Chamaedaphne calyculata (in Eurasia/NW North American and NE North American haplogroup), b Linnaea borealis ssp. borealis, and c Pedicularis sceptrum-carolinum ssp. sceptrum-carolinum. Vm Vaccinium macrocarpon, Ac Abelia corymbosa, Ps-c Pedicularis sylvatica as the outgroup species. Circles with black and orange hatching indicate populations/cpDNA and ITS haplotypes projected to be lost in the future (2070) under RCP 4.5, 6.0, and 8.5 climate projections. ITS markers were polymorphic only for Chamaedaphne calyculata. Gray scale refers to logistic probability (0–1) of taxon occurrence, darkening with increasing climate suitability
In L. b ssp. borealis, six cpDNA haplotypes were identified in the dataset (Table 1, Supplementary information Table S3). Haplotype A had the widest distribution and was found in formerly glaciated and non-glaciated areas in Europe in 20 populations. The distribution of the five other haplotypes across the range revealed higher haplotype diversity in the south: haplotype B was detected in two populations in Northern and Southern Europe (Alps), haplotype C in Southern Europe and the Alps, and three unique haplotypes D, E, and F in the Alps and Carpathians (Fig. 2b). All the haplotypes formed one group with 100% posterior probability (Fig. 2b).
Four cpDNA haplotypes were identified in P. s-c ssp. sceptrum-carolinum (A–E, Fig. 2c, Table 1, Supplementary information Table S3). Haplotype A was the most widespread, present in 13 populations in formerly glaciated areas in Europe and one Carpathian population in a non-glaciated area (Fig. 2c). Haplotype B was restricted to non-glaciated territory of South Asia and one northern population of a formerly glaciated European range (Fig. 2c). One cpDNA haplotype (C) was found exclusively in the southern Carpathians, another one (D) in East Asia, and haplotype E in Northern Europe (Fig. 2c). The Bayesian tree of cpDNA haplotypes built by BEAST revealed two well-supported groups: Eurasian (77% bootstrap support) and Carpathian-East Asian (85% bootstrap support, Fig. 2c).
The predicted future of phylogeographic structure varies under different climate models (Table 1), but in three scenarios, the unique cpDNA haplotypes in each studied taxon will be lost in the future, while the common ones will be maintained. We forecasted that in C. calyculata, genetic erosion might involve two unique haplotypes (C and D) under RCP 4.5–8.5 in the NE North American group. The slight loss of haplotypes and decrease of h values were assessed from the ITS data in the Eurasian/NW North American group under RCP 4.5–8.5. In the NE North American group, potential loss of ITS haplotypes was ca. 22% under RCP 4.5 and 6.0, but the loss of ITS haplotype ca. 44% and h ca. 47% were considerably higher under RCP 8.5 for the future than the present (Table 1).
In L. b ssp. borealis, the loss of two out of six cpDNA haplotypes, clustered in one haplogroup, was predicted for 2070 in all RCPs. The recalculated h and π values for the populations remaining in climatically suitable areas in the future are 11–23% lower (Table 1). In P. s-c ssp. sceptrum-carolinum, the distribution of phylogeographic diversity may lead to the loss of two cpDNA haplotypes (C, D), which form one distinct haplogroup, while h becomes ca. 48% lower than the current genetic diversity (Table 1).
AFLP analysis
A total of 220–343 loci were resolved per plant taxon, with error rates of 2.7–3.1%. The three plant taxa differed in genetic diversity levels. After adjustment for uneven sample size by rarefaction, C. calyculata showed relatively low H values (0.084 for Eurasian/NW North American, 0.091 for NE North American group), similar to L. b ssp. borealis (0.089); the value for P. s-c ssp. sceptrum-carolinum (0.156) was double that that of the other two. The DW values for L. b ssp. borealis (1.62), P. s-c ssp. sceptrum-carolinum (1.46), and C. calyculata for the NE North American group (1.50) were quite similar; in C. calyculata for the Eurasian/NW North American group, the values were two times lower (0.73) than of the others. When the level of suitability for taxa persistence was applied to the estimated H values for the taxa under future climate change, the respective values for the two groups changed slightly in C. calyculata (0.083 in all three scenarios as well as 0.086 in RCP 4.5 and 6.0 and 0.099 in RCP 8.5, respectively; Table 1); in L. b ssp. borealis, the values were similar to the current level of gene diversity (0.087 in RCP 4.5–8.5; Table 1), and in P. s-c ssp. sceptrum-carolinum, they were 0.153 in three scenarios. In the other two circumboreal taxa, the DW values tended to decrease in the future: in the Eurasian/NW North American group of C. calyculata, the value was likewise ca. 12% lower (0.64, Table 1) and in the NE North American group ca. 23% lower (1.15, Table 1) for the future than for the present. In L. b ssp. borealis, the DW was 1.55 in RCP 4.5 and 1.67 in RCP 6.0 and 8.5. In P. s-c ssp. sceptrum-carolinum, the DW value was ca. 8% lower than for the present (Table 1). The AFLP data provided strong evidence for a correlation between longitude and DW values in C. calyculata (r = 0.50, p < 0.01, Supplementary information Fig. S3a) as well as latitude and DW values in L. b ssp. borealis only (r = − 0.567, p < 0.01, Supplementary information Fig. S3b). In P. s-c ssp. sceptrum-carolinum, there was a significant association between H and BIO1 (annual mean temperature, r = 0.51, p < 0.05, Supplementary information Fig. S3c). In three circumboreal taxa, the relationship between estimated loss of genetic diversity and loss of geographical range was nonlinear; and the 95% confidence interval (CI) revealed that uncertainties referring to loss of genetic and geographical range were low (Supplementary information Fig. S3).
The number of clusters generated in POPS in the model of current distribution differed among the studied three plants. To obtain more detailed information about their genetic structure, we considered the cluster analysis in two separate phylogeographic groups for C. calyculata. In the Eurasian/NW North American group, the DIC curve plateaued at K max = 5; in the NE North American group, K max was at 3 (Supplementary information Fig. S4a). In the Eurasian/NW North American group, three genetic pools dominated in NW North America, and three others in Eurasia (Fig. 3a). The specimens from six populations in this group represented a strong mixture of diverse genetic backgrounds. In the NE North American group, one distinct genetic pool was identified in southeastern North America; the others were present within the remaining populations. Admixture was observed in only two populations: one from the southern edge of the range and the other from eastern North America (Fig. 3a); the respective correlations between membership coefficient and predicted membership coefficient, r mp, based on current bioclimatic variables, were 0.83 and 0.98. DIC analysis showed K max = 2 in L. b ssp. borealis (Supplementary information Fig. S4b; r mp = 0.82); the two clusters did not reflect a clear geographic structure, and almost all individuals represented a mixture of two diverse genetic backgrounds (Fig. 3b). Four genetic pools were identified (K max = 4) in P. s-c ssp. sceptrum-carolinum (r mp = 0.88, Supplementary information Fig. S4c). One cluster dominated in one Asian population, and the rest of the European and Asian P. s-c ssp. sceptrum-carolinum populations comprised individuals with various levels of admixture of three other gene pools (Fig. 3c).
Present-day and future population genetic structure (2070) simulated under RCP 4.5, 6.0, and 8.5 climate scenarios inferred by POPS for Chamaedaphne calyculata (presented for Eurasian/NW North American haplogroup, Kmax = 5, and for NE North American haplogroup, Kmax = 3, a), Linnaea borealis ssp. borealis (Kmax = 2, b), and Pedicularis sceptrum-carolinum ssp. sceptrum-carolinum (Kmax = 4, c). Colors represent the genetic groups identified, pie chart shows the proportion of gene pool in each population. Gray scale refers to logistic probability (0–1) of taxon occurrence, darkening with increasing climate suitability
The POPS ancestry distribution model provided projections of genetic structure in circumboreal plant species under climate changes in 2070. The scenario of changes showed some dynamism. The predicted genetic structure differed between two phylogeographic groups in C. calyculata: Eurasia/NW North America still would have five clusters in the future but with an increased share of one cluster (green) in the Central European and East Asian populations. In the NE North American C. calyculata populations, one cluster was no longer present, and another cluster (orange) dominated the majority of populations (Fig. 3a). It was similar in L. b ssp. borealis: almost all populations were clustered into one homogenous gene pool (Fig. 3b). The future pattern of population genetic structure was slightly different in P. s-c ssp. sceptrum-carolinum, where four genetic pools would still exist, but in case of the more severe RCP 8.5 scenario, a few populations were characterized by the homogenous gene pools similar to the other studied circumboreal plants (Fig. 3c).
Discussion
Current and future predictions of the geographic and genetic diversity pattern of circumboreal plants
Our investigation suggests that the geographic range and genetic diversity patterns of circumboreal plant taxa will respond in different ways under future climate warming. The three circumboreal plants appear to lose their geographic ranges in the future (2070) under the three scenarios (RPC 4.5, 6.0, and 8.5). The range may withdraw 35–52% under the lowest-emission RCP 4.5 scenario and 56–69% under more severe RPC 8.5 scenario, only slightly compensated by predicted range gains of 18–33% in three scenarios. However, those numbers should be used with caution, since mean climatically suitable areas in our study differed up to 50% from average, depending on the climate forecasting model. In our approach, we have averaged six different GCMs to account for uncertainty of future climatic conditions and mitigate prediction of potential range changes. We believe this should be a common approach, when extrapolating similar studies to future conditions. We estimated that the distribution of suitable climatic conditions for these plants will shift by ca. 8° latitude to the north in Eurasia and North America. This is in accord with work on Arctic plants predicting that at the end of the century loss of geographic range will generally be higher than the range gain (Alsos et al. 2012), but does not correspond to the twofold lower values given for Mediterranean-temperate tree species (Temunovic et al. 2013) and temperate tree species under different emission scenarios (Morin and Viner et Chuine 2008). In our different climatic scenarios, the (highly isolated) Central European populations of the three circumboreal plants we studied will become critically endangered or even extinct in the future. On the other hand, the populations in southern Fennoscandia will rather not be at the risk of extinction suggested by Thuiller et al. (2005), as they may exist in areas of predicted suitable climate. In the Asian distribution of the three circumboreal taxa, the potential main area of range loss is in the central and northeastern parts of the continent in all three climate scenarios.
Predicting the future geographic range of circumboreal plants adapted to specific habitats such as Sphagnum-dominated peatlands (C. calyculata) or moist meadows and peat bogs (P. s-c ssp. sceptrum-carolinum) is a complicated task. These habitats could be lost faster than their creation (Galloway et al. 1995), even if climatically suitable areas for these taxa still exist in the current and future distributions. Thuiller et al. (2005) focused on another problem of plant diversity under climate change. Many European boreal regions may not only lose species but also gain species from southern parts of Europe. This would change the floristic composition of communities and foster competition between newly established and resident plant species.
Recent estimates of the rate of global climate change suggest that species may not be able to shift their ranges fast enough to track suitable conditions (Loarie et al. 2009), and that species survival depends not only on neutral genetic diversity but on factors such as phenotypic plasticity, adaptive capacity, mutation rate and/or introgression, founder events, etc. (Hoffmann and Sgró 2011). If there is an evolutionary-based perspective to the survival of populations in climatically suitable areas, these populations should be viewed especially as a matter of concern in conservation biology because it allows species to occupy a wide range of environmental niches, especially ones with broad geographic distributions. Our conclusions on the putative future genetic diversity patterns of circumboreal plants, as in most surveys, are based on surrogate neutral genetic diversity, and they rest on assumptions including no long distance dispersal and/or niche conservatism. With ENM-based methods, it is difficult to introduce interspecific biotic interaction, dispersal, and migration scenarios into projections of future distribution. We did not incorporate such features in the ENMs, leading to overestimation of the future plant distributions as well as to incongruity between the empirical and modeled spatial genetic structures.
Despite these uncertainties and the positing of assumptions that can rather increase the putative loss of genetic diversity in species (Alsos et al. 2012), our results demonstrate the usefulness of combining ENMs and molecular genetic analyses to forecast spatial patterns of genetic diversity. The changes in the genetic diversity patterns of these circumboreal plants depended on the type of molecular marker and patterns of genetic diversity distribution. This fact was also supported in a few other surveys, i.e., Collevatti et al. (2011), Alsos et al. (2012), Jay et al. (2012), and Temunovic et al. (2013). In the case of circumboreal plants, the expected future loss of geographic range was not necessary accompanied by a marked reduction of genetic diversity, but it has a strong effect on allelic richness, even if genetic diversity was quite uniformly distributed. Indeed, the loss of rare alleles without any substantial loss of genetic diversity is predicted after population size reduction and genetic drift will tend to make rare alleles to disappear more rapidly than common alleles. Nei’s gene diversity is weakly dependent on the loss of rare alleles, whereas allelic richness is sensitive to the number of alleles whatever their prevalence (Allendorf 1986). Our study showed that climatically stable areas cannot be the exclusive, decisive factor determining the patterns of genetic structure and diversity in circumboreal taxa as was suggested by Ortego et al. (2014). Therefore, when we used the chloroplast and nuclear markers, genetic diversity was shown either to potentially remain at present-day levels or decline, while rare and unique cpDNA and ITS haplotypes in C. calyculata and L. b ssp. borealis probably will undergo significant loss. C. calyculata showed a loss of geographic range (35–56% in three RCP scenarios) and erosion of genetic diversity using nuclear (ca. 12–23%) and chloroplast (loss of two of seven cpDNA haplotypes that formed a distinct haplogroup) markers in 2070; we did observe depauperation of gene pools. A different scenario was predicted for P. s-c ssp. sceptrum-carolinum: we found slight but not significant differences between the southern and northern populations within their European geographic range, but loss of Southern European populations of P. s-c ssp. sceptrum-carolinum could cause a ca. 8% reduction of its genetic diversity in the future.
Our work also indicated that the number of gene pools and their degree of admixture in the populations throughout the distribution range was the second most important factor determining future genetic diversity patterns under climate change. Interestingly, the similarity of current and predicted (2070) genetic diversity in C. calyculata and L. b ssp. borealis estimated using AFLP markers did not match the pattern for population genetic structure. The homogeneity of the genetic background—the lack of admixture and the domination of one gene pool in most populations throughout the ranges of these two plants—becomes more pronounced in the future. In this case, the ancestry distribution model combined with geographic distribution of populations and environmental data (Jay et al. 2012) assumes gene flow as the main component in the response of plant populations to environmental modifications and favors migration toward similar environments. Therefore, this model showed that resident genes can move to suitable places in a way that conserves the realized niche of a given genetic cluster. Only one study of future genetic diversity has reported severe erosion of a taxon’s gene pool and decreased admixture in its populations. A simulation of the genetic structure of the Mediterranean tree Fraxinus angustifolia in a future climate scenario predicted that one cluster would no longer exist (Temunovic et al. 2013). One or two gene pools that are dominant in populations in present-day suitable areas may replace other gene pools occurring at low frequency in other populations still existing in the future. Predicting which populations will be lost, and therefore which components of genetic diversity and the gene pool will be lost, is an important task.
The lack of a particular dispersal adaptation in the small seeds of the investigated circumboreal plants may not affect their colonization ability. The moderate level of genetic differentiation estimated for these taxa (F ST = 0.173–0.289) was probably the consequence of short-distance and random long-distance dispersal, the ability to colonize specific habitats, and rapid postglacial range expansion in the Northern Hemisphere (Wróblewska 2012, 2013a, b); this suggests that the lack of a dispersal adaptation will not be a major factor affecting loss of genetic diversity in these plants. For many clonal plants, recruitment from seeds is important during initial colonization (Bazzaz 1996). The relatively low genetic differentiation among populations of circumboreal taxa might be explained by the finding of Aguinagalde et al. (2005) that species of the boreal-temperate group were probably geographically less restricted and isolated during glacial maxima, which would lead to stronger dynamics of population establishment and extinction, as well as greater seed-mediated gene flow between existing populations.
Identification of past climatic refugia and future macrorefugia
Phyloclimatic modeling that uses species distribution and phylogeographic models indicates refugial areas that are spatially and temporally more explicit than the areas given in the outcomes of either model alone (Waltari et al. 2007; Carnaval et al. 2009; Keppel et al. 2012). Pauls et al. (2013) and Gavin et al. (2014) expanded this approach to include fossil data. This new integrative approach better identified refugia and postglacial colonization routes in Fagus sylvatica, Pseudotsuga menziesii, and Picea crassifolia (Gavin et al. 2014).
The origin of our three studied circumboreal taxa has not been understood well. They are all insect-pollinated taxa, and palynological data from peat layers are sparse. In only one case, C. calyculata macrofossil records (seeds and leaves) were quite well investigated throughout the former distribution. Macrofossils of this species from the Late Tertiary have been found in non-glaciated regions of Alaska (Matthews and Ovenden 1990) and in northern and northeastern Siberia (Nikitin 2006), from the end of the Wisconsin interglacial in Nova Scotia (Mott et al. 1982), and in the Early Würm in Siberia (Laukhin et al. 2007). Other studies have reported only Early, Middle, or Late Holocene macrofossils from North America, Europe, and Asia (Bhiry and Filion 1996; Booth and Jackson 2003; Booth et al. 2004; Sannel and Kuhry 2008; Novenko et al. 2009). The pollen and macrofossil records of the other two taxa, L. b ssp. borealis and P. s-c ssp. sceptrum-carolinum, described their occurrence in the Holocene in Eurasia (Rosén et al. 2001; Anderson et al. 2006; Mol et al. 2006; Hájková et al. 2012). The ENMs predicted a far more widespread past distribution for each circumboreal taxon under climate simulations. In the case of C. calyculata, the combination of cpDNA and ITS data with paleo-ENMs and macrofossil records resolved the putative refugial areas in Beringia and southern North America more precisely (Wróblewska 2012). An integrated reconstruction of refugia during the LGM using the molecular data from chloroplast and nuclear markers along with paleo-ENMs showed that L. b ssp. borealis may have persisted in East or Central Asia (Tibetan Plateau) and in the Alps during the LGM (Wróblewska 2013b). The paleoclimate models indicate that the past distribution of circumboreal taxa, particularly C. calyculata and L. b ssp. borealis during the late glacial, partially overlaps the distribution of boreal vegetation in Central, Southern and Eastern Asia as well as Central and Southern Europe (Allen et al. 2010). Identifying refugia is more complicated in P. s-c ssp. sceptrum-carolinum due to its stronger fragmentation in Europe and its disjunctive current distribution in Asia. The Bayesian approach and chloroplast data do not precisely reveal the areas where this taxon could have persisted during the LGM, probably because the refugial populations were lost due to contraction of the range. P. s-c ssp. sceptrum-carolinum refugia are found from complementary research such as paleo-ENMs as well as floristic and taxonomic diversity studies of the genus Pedicularis which show their source of expansion to be rather in East Asia (Hong 1983; Tkach et al. 2014). The macrorefugia for the studied circumboreal plants, meaning the putative overlap of current and future climatically suitable areas, are predicted to be restricted to northern parts of Eurasia between ca. 53–64° latitude and North America between ca. 48–59°, reaching the arctic zone. Our results suggest overlapping of 25–50% between the present and predicted distribution of these taxa; this will affect the survival of several studied populations and their future genetic resources. Intrinsic dispersal limitation seems more important for restricted-range species. It can be also argued that restricted-range species may be responding to climate gradients on scales smaller than those of widespread species (Hawkins and Diniz-Filho 2006).
Conclusions
Climate change is considered as one of the factors influencing genetic diversity of plant species. In this study, the important question is how species’ genetic diversity will change when species are moving as result of climate change. Most studies usually show predictions that global climate change may lead to the loss of future species’ ranges ignoring intraspecific future genetic diversity. In the present study, an interdisciplinary approach integrating both future climate change scenarios (in 2070) under three representative concentration pathways (4.5, 6.0, and 8.5 RCPs) and genetic diversity data reveals taxon-specific responses to future climate change. The current geographic distribution of the three circumboreal plants studied here would be strongly lost in the future and only slightly compensated by a predicted range gain. Simultaneously, the overall genetic diversity of studied plants could remain similar to or lower than the present level, while unique cpDNA and ITS haplotypes will likely be lost at the taxon level. Additionally, genetic structure based on spatial Bayesian clustering described by Jay (2011) clearly reveals the homogeneity of the genetic background due to a lack of admixture and domination of one gene pool in most populations of these plants in the future. There is no doubt that a more multidisciplinary approach should be applied, coupling ENMs and genetic studies to better understand the processes of gene flow, genetic drift, and adaptation at the species level in the future. Moreover, more comprehensive studies using several molecular markers (e.g., chloroplast and nuclear DNA) are needed to interpret with greater precision perspectives of species’ survival in the future, which a single molecular marker would not necessarily reveal.
References
Abbott RJ, Comes HP (2004) Evolution in the Arctic: a phylogeographic analysis of the circumarctic plant Saxifraga oppositifolia (purple saxifrage). New Phytol 161:211–224. doi:10.1046/j.1469-8137.2003.00953.x
Aguinagalde I, Hampe A, Mohanty A, Pedro Martin J, Duminil J, Petit RJ (2005) Effects of life-history traits and species distribution on genetic structure at maternally inherited markers in European trees and shrubs. J Biogeogr 32:329–339. doi:10.1111/j.1365-2699.2004.01178.x
Allen JRM, Hickler T, Singarayer JS, Sykes MT, Valdes PJ, Huntley B (2010) Last glacial vegetation of northern Eurasia. Quat Sci Rev 29:2604–2618. doi:10.1016/j.quascirev.2010.05.031
Allendorf FW (1986) Genetic drift and the loss of alleles versus heterozygosity. Zoo Biol 5:181–190. doi:10.1002/zoo.1430050212
Alsos IG, Alm T, Normand S, Brochmann C (2009) Past and future range shifts and loss of diversity in dwarf willow (Salix herbacea L.) inferred from genetics fossils and modelling. Glob Ecol Biogeogr 18:223–239. doi:10.1111/j.1466-8238.2008.00439.x
Alsos IG, Ehrich D, Thuiller W, Eidesen PB, Tribsch A, Schönswetter P, Lagaye C, Taberlet P, Brochmann C (2012) Genetic consequences of climate change for northern plants. Proc R Soc Lond B Biol Sci 279:2042–2051. doi:10.1098/rspb.2011.2363
Anderson RS, Hallett DJ, Berg E, Jass RB, Toney JL, de Fontaine CS, DeVolder A (2006) Holocene development of boreal forests and fire regimes on the Kenai lowlands of Alaska. The Holocene 16:791–803. doi:10.1191/0959683606hol966rp
Ashcroft MB (2010) Identifying refugia from climate change. J Biogeogr 37:1407–1413. doi:10.1111/j.1365-2699.2010.02300.x
Barrett SCH, Husband BC (1990) Genetics of plant migration and colonization. In: Brown AHD, Clegg MT, Kahler AL, Weir BS (eds) Plant population genetics breeding and genetic resources. Sinauer Associates Inc., Massachusetts, pp 254–277
Bazzaz FA (1996) Plants in changing environments. Linking physiological population and community ecology. Cambridge University Press, Cambridge
Beatty GE, Provan J (2010) Refugial persistence and postglacial recolonization of North America by the cold-tolerant herbaceous plant Orthilia secunda. Mol Ecol 19:5009–5021. doi:10.1111/j.1365-294x
Beatty GE, Provan J (2011) Phylogeographic analysis of north American populations of the parasitic herbaceous plant Monotropa hypopitys L. reveals a complex history of range expansion from multiple late glacial refugia. J Biogeogr 38:1585–1599. doi:10.1111/jbi.12371
Betts RA, Golding N, Gonzalez P, Gornall J, Kahana R, Kay G, Mitchell L, Wiltshire A (2015) Climate and land use change impacts on global terrestrial ecosystems and river flows in the HadGEM2-ES earth system model using the representative concentration pathways. Biogeosciences 12:1317–1338. doi:10.5194/bg-12-1317-2015
Bhagwat SA, Wilis JK (2008) Species persistence in northerly glacial refugia of Europe: a matter of chance or biogeographical traits? J Biogeogr 35:464–482. doi:10.1111/j.1365-2699.2007.01861.x
Bhiry N, Filion L (1996) Holocene plant succession in a dune-swale environment of southern Quebec: a macrofossil analysis. Ecoscience 3:330–334
Bizoux JP, Mahy G (2007) Within-population genetic structure and clonal diversity of a threatened endemic metallophyte Viola calaminaria (Violaceae). Am J Bot 94:887–895. doi:10.3732/ajb.94.5.887
Booth RK, Jackson ST (2003) A high-resolution record of late Holocene moisture variability from a Michigan raised bog. The Holocene 13:865–878. doi:10.1007/s00334-014-0466-y
Booth RK, Jackson ST, Gray CED (2004) Paleoecology and high-resolution paleohydrology of kettle-hole peatland in upper Michigan. Quat Res 61:1–13. doi:10.1016/j.yqres.2003.07.013
Carnaval A, Hickerson MJ, Haddad CFB, Rodrigues MT, Moritz C (2009) Stability predicts genetic diversity in the Brazilian Atlantic Forest Hotspot. Science 323:785–789. doi:10.1016/j.ympev.2009.09.016
Collevatti RG, Nabout JC, Diniz-Filho JAF (2011) Range shift and loss of genetic diversity under climate change in Caryocar brasiliense a Neotropical tree species. Tree Genet Genomes 7:1237–1247. doi:10.1007/s11295-011-0409-z
Drummond AJ, Suchard MA, Xie D, Rambaut A (2012) Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol 29(8):1969–1973. doi:10.1093/molbev/mss075
Dufresne J-L, Foujols M-A, Denvil S, Caubel A, Marti O, Aumont O, Balkanski Y, Bekki S, Bellenger H, Benshila R, Bony S, Bopp L, Braconnot P, Brockmann P, Cadule P, Cheruy F, Codron F, Cozic A, Cugnet D, de Noblet N, Duvel J-P, Ethà C, Fairhead L, Fichefet T, Flavoni S, Friedlingstein P, Grandpeix J-Y, Guez L, Guilyardi E, Hauglustaine D, Hourdin F, Idelkadi A, Ghattas J, Joussaume S, Kageyama M, Krinner G, Labetoulle S, Lahellec A, Lefebvre M-P, Lefevre F, Levy C, Li ZX, Lloyd J, Lott F, Madec G, Mancip M, Marchand M, Masson S, Meurdesoif Y, Mignot J, Musat I, Parouty S, Polcher J, Rio C, Schulz M, Swingedouw D, Szopa S, Talandier C, Terray P, Viovy N, Vuichard N (2013) Climate change projections using the IPSL-CM5 earth system model: from CMIP3 to CMIP5. Clim Dyn 40:2123–2165. doi:10.1007/s00382-012-1636-1
Ehrich D (2006) AFLPdat: a collection of R functions for convenient handling of AFLP data. Mol Ecol Notes 6:603–604. doi:10.1111/j.1471-8286.2006.01380.x
Ehrich D, Alsos IG, Brochmann C (2008) Where did the northern peatland species survive the dry glacials? Cloudberry (Rubus chamaemorus) as an example. J Biogeogr 35:801–814. doi:10.1111/j.1365-2699.2007.01864.X
Eidesen PB, Alsos IG, Popp M, Řd S, Brochmann C (2007) Nuclear versus plastid data: complex Pleistocene history of a circumpolar key species. Mol Ecol 16:3902–3925. doi:10.1111/j.1365-294X.2007.03425.x
Eidesen PB, Ehrich D, Bakkestuen V, Alsos IG, Gilg O, Taberlet P, Brochmann C (2013) Genetic roadmap of the Arctic: plant dispersal highways traffic barriers and capitals of diversity. New Phytol 200:898–910. doi:10.1111/nph.12412
Excoffier L, Laval G, Schneider S (2005) ARLEQUIN (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinformatics Online 1:47–50
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Mol Ecol Notes 7:574–578. doi:10.1111/j.1471-8286.2007.01758.x
Galloway JN, Schlesinger WH, Levy H, Michaels A, Schnoor JL (1995) Nitrogen fixation: Anthropogenic enhancement-environmental response. Glob Biogeochem Cycles 9:235–252. doi:10.1023/A:1006247623877
Gavin DG, Fitzpatrick MC, Gugger PF, Heath KD, Rodríguez-Sánchez F, Solomon Z (2014) Climate refugia: joint inference from fossil records species distribution models and phylogeography. New Phytol 204:37–54. doi:10.1111/nph.12929
Giovanelli JGR, Ferreira de Siqueira M, Haddad CFB, Alexandrino J (2010) Modeling a spatially restricted distribution in the Neotropics: how the size of calibration area affects the performance of five presence-only methods. Ecol Model 221:215–224. doi:10.1016/j.ecolmodel.2009.10.009
Gitzendanner MA, Soltis S (2000) Patterns of genetic variation in rare and widespread plant congeners. Amer J bot 87:783 –792. doi:10.5735/085.047.0605
Guisan A, Thuiller W (2005) Predicting species distribution: offering more than simple habitat models. Ecol Lett 8:993–1009. doi:10.1111/j.1461-0248.2005.00792.x
Hájková P, Grootjans AB, Lamentowicz M, Rybničková E, Madaras M, Opravilová V, Michaelis H, Hájek M, Joosten H, Wolejko L (2012) How a Sphagnum fuscum-dominated bog changed into a calcareous fen: the unique Holocene history of a Slovak spring-fed mire. J Quat Sci 27(3):233–243. doi:10.1002/jqs.1534
Hamrick JL, Godt MJW (1989) Allozyme diversity in plant species. In: MT BAHDC, Kahler AL, Weir BS (eds) Plant population genetics breeding and genetic resources. Sinauer Associates, Sunderland
Hawkins BA, Diniz-Filho JAF (2006) Beyond Rapoport’s rule: evaluating range size patterns of New World birds in a two-dimensional framework. Global Ecol Biogeogr 15:461–469. doi:10.1111/j.1466-822X.2006.00243.x
Hoffmann AA, Sgró CM (2011) Climate change and evolutionary adaptation. Nature 470:479–485. doi:10.1038/nature09670
Hong D (1983) The distribution of Scrophulariaceae in the Holarctic with special reference to the floristic relationships between eastern Asia and eastern North America. Ann Mo Bot Gard 70:701–713. doi:10.2307/2398985
Hultén E, Fries M (1986) Atlas of north European vascular plants. 1. Koeltz Scientifi C books Königstein
Jay F (2011) POPS: prediction of population genetic structure-program documentation and tutorial. University Joseph Fourier Grenoble France. Available from http://membres-timc.imag.fr/Olivier.Francois/pops.html
Jay F, Manel S, Alvarez N, Durand EY, Thuiller W, Holderegger R, Taberlet P, François O (2012) Forecasting changes in population genetic structure of alpine plants in response to global warming. Mol Ecol 21:2354–2368. doi:10.1111/j.1365-294X.2012.05541.x
Jørgensen MH, Elven R, Tribsch A, Gabrielsen TM, Stedje B, Brochmann C (2006) Taxonomy and evolutionary relationships in the Saxifraga rivularis Complex. Syst Bot 31:702–729. doi:10.1600/036364406779695988
Jump A, Peñuelas J (2006) Genetic effects of chronic habitat fragmentation in a wind-pollinated tree. Proc Natl Acad Sci U S A 103:8096–8100. doi:10.1073/pnas.0510127103
Jump AS, Mátyás C, Peñuelas J (2009) The altitude-for-latitude disparity in the range retractions of woody species. Trends Ecol Evol 24:694–701. doi:10.1016/j.tree.2009.06.007
Keller SR, Olson MS, Silim S, Schroeder W, Tiffin P (2010) Genomic diversity population structure and migration following rapid range expansion in the balsam poplar Populus balsamifera. Mol Ecol 19:1212–1226. doi:10.1093/molbev/mss121
Keppel G, Van Niel KP, Wardell-Johnson GW, Yates CJ, Byrne M, Mucina L, Schut AGT, Hopper SD, Franklin SE (2012) Refugia: identifying and understanding safe havens for biodiversity under climate change. Glob Ecol Biogeogr 21:393–404. doi:10.1111/j.1466-8238.2011.00686.x
Laukhin SA, Arslanov KA, Maksimov FE, Kuznetsov VY (2007) The first early Interstadial of Zirianian traces (early Würm) glaciation in Siberia: U/Th date and palaeobotanical data. Geologija 59:47–58
Liu C, Pam M, Dawson TP, Pearson RG (2005) Selecting thresholds of occurrence in the prediction of species distributions. Ecography 28:385–393. doi:10.1111/j.0906-7590.2005.03957.x
Loarie SR, Duffy PB, Hamilton H, Asner GP, Field CB, Ackerly DD (2009) The velocity of climate change. Nature 462:1052–1055. doi:10.1038/nature08649
Lobo JM, Jiménez-Valverdel A, Real R (2008) AUC: a misleading measure of the performance of predictive distribution models. Glob Ecol Biogeogr 17:145–151. doi:10.1111/j.1466-8238.2007.00358.x
Loveless MD, Hamrick JL (1984) Ecological determinants of genetic structure in plant populations. Annu Rev Ecol Evol Syst 15:65–95. doi:10.1146/annurev.es.15.110184.000433
Masui T, Matsumoto K, Hijioka Y, Kinoshita T, Nozawa T, Ishiwatari S, Kato E, Shukla PR, Yamagata Y, Kainuma M (2011) An emission pathway for stabilization at 6 Wm-2 radiative forcing. Clim Chang 109:59–76. doi:10.1007/s10584-011-0150-5
Matthews JV, Ovenden LE (1990) Late tertiary plant macrofossils from localities in Arctic/subarctic North America: a review of the data. Arctic 43:364–392. DOI: 10.14430/arctic1631
Meusel H, Jäger E, Rauschert S, Weinert E (1978) Vergleichende Chorologie der zentraleuropäischen Flora. Fischer, Jena
Mitka J, Boroń J, Wróblewska A, Bąba W (2015) AFLP analysis reveals infraspecific phylogenetic relationships and population genetic structure of two species of Aconitum in Central Europe. Acta Soc Bot Pol 84(2):267–276. doi:10.5586/asbp.2015.012
Mol D, Tikhonovb A, van der Plichtc J, Kahlked R-D, Debruynee R, van Geelf B, van Reenenf G, Palsg JP, de Marliaveh C, Reumer JWF (2006) Results of the CERPOLEX/Mammuthus expeditions on the Taimyr peninsula Arctic Siberia Russian Federation. Quat Int 142–143:186–202. doi:10.1016/j.quaint.2005.03.016
Morin X, Viner et Chuine D (2008) Tree species range shifts at a continental scale: new predictive insights from a process-based model. J Ecol 96:784–794. doi:10.1111/j.1365-2745.2008.01369.x
Mott RJ, Anderson TW, Matthews JV (1982) Pollen and macrofossil study of an interglacial deposit in Nova Scotia. Géog Phys Quatern 36:197–208
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York. doi:10.1002/ajpa.1330750317
Nenzén HK, Araújo MB (2011) Choice of threshold alters projections of species range shifts under climate change. Ecol Model 222:3346–3354. doi:10.1016/j.ecolmodel.2011.07.011
Nikitin VP (2006) Paleogene and Neogene strata in North-eastern Asia: paleocarpological background. Russ Geol Geophys 48:675–682. doi:10.1016/j.rgg.2006.06.002
Normand S, Treier UA, Randin C, Vittoz P, Guisan A, Svenning J-C (2009) Importance of abiotic stress as a range-limit determinant for European plants: insights from species responses to climatic gradients. Glob Ecol Biogeogr 18:437–449. doi:10.1111/j.1466-8238.2009.00451.x
Novenko EY, Volkova EM, Nosova NB, Zuganova IS (2009) Late glacial and Holocene landscape dynamics in the southern taiga zone of the east European plain according to pollen and macrofossil records from the central Forest state reserve. Quat Int 207:93–103. doi:10.1016/j.quaint.2008.12.006
Nybom H (2004) Comparison of different nuclear DNA markers for estimating intraspecific genetic diversity in plants. Mol Ecol 13:1143–1155. doi:10.1111/j.1365-294X.2004.02141.x
Oksanen J, Blanchet FG, Kindt R, Legendre P, O’Hara RB, Simpson GL (2015) Vegan: community ecology package. R package version 1.17–4. http://cran.r-project.org/
Ortego J, Gugger PF, Riordan EC, Sork VL (2014) Influence of climatic niche suitability and geographic overlap on hybridization patterns among southern Californian oaks. J Biogeogr 41:1895–1908. doi:10.1111/jbi.12334
Parmesan C (2006) Ecological and evolutionary responses to recent climate change. Annu Rev Ecol Evol Syst 37:637–669. doi:10.1146/annurev.ecolsys.37.091305.110100
Pauls C, Nowak M, Bálint M, Pfenninger C (2013) The impact of global climate change on genetic diversity within populations and species. Mol Ecol 22:925–946. doi:10.1111/mec.12152
Phillips SJ, Anderson RP, Schapire RE (2006) Maximum entropy modeling of species geographic distributions. Ecol Model 190:231–259. doi:10.1016/j.ecolmodel.2005.03.026
Posada D (2008) jModelTest: phylogenetic model averaging. Mol Biol Evol 25:1253–1256. doi:10.1093/molbev/msn083
Rambaut A (2010) FIGTREE 1.3.1. Available at: http://tree.bio.ed.ac.uk/software/figtree/
Riahi K (2011) RCP8.5 – a scenario of comparatively high greenhouse gas emissions. Clim Chang 109:33–57. doi:10.1007/s10584-011-0149-y
Rosén P, Segerström U, Eriksson L, Renberg I, Birks HJB (2001) Holocene climatic change reconstructed from diatoms chironomids pollen and near-infrared spectroscopy at an alpine lake (Sjuodjijaure) in northern Sweden. The Holocene 11:551–562. doi:10.1191/095968301680223503
Sannel ABK, Kuhry P (2008) Long-term stability of permafrost in subarctic peat plateaus west-central Canada. The Holocene 18:589–601. doi:10.1177/0959683614538073
Schönswetter P, Tribsch A (2005) Vicariance and dispersal in the alpine perennial Bupleurum stellatum L. (Apiaceae). Taxon 54:725–732. doi:10.2307/25065429
Schönswetter P, Suda J, Popp M, Weiss-Schneeweiss H, Brochmann C (2007) Circumpolar phylogeography of Juncus biglumis (Juncaceae) inferred from AFLP fingerprints cpDNA sequences nuclear DNA content and chromosome numbers. Mol Phylogenet Evol 42:92–103. doi:10.1016/j.ympev.2006.06.016
Steane DA, Potts BM, McLean E, Prober SM, Stock WD, Vaillancourt RE, Byrne M (2014) Genome-wide scans detect adaptation to aridity in a widespread forest tree species. Mol Ecol 23:2500–2513. doi:10.1111/mec.12751
Svenning JC, Fløjgaard C, Marske KA, Nógues-Bravo D, Normand S (2011) Applications of species distribution modeling to paleobiology. Quat Sci Rev 30:2930–2947. doi:10.1016/j.quascirev.2011.06.012
Temunovic M, Frascaria-Lacoste N, Franjic J, Satovic Z, Fernandez-Manjarres JF (2013) Identifying refugia from climate change using coupled ecological and genetic data in a transitional Mediterranean-temperate tree species. Mol Ecol 22(8):2128–2142. doi:10.1111/mec.12252
Thomassen HA, Buermann BM, Graham CH, Cameron SE, Schneider CJ, Pollinger JP (2010) Modeling environmentally associated morphological and genetic variation in a rainforest bird and its application to conservation prioritization. Evol Appl 3:1–16. doi:10.1111/j.1752-4571.2009.00093.x
Thomson C, Smith K, Volke P, Delgado A, Wise B-L, Clarke E (2011) RCP4.5: a pathway for stabilization of Radiative forcing by 2100. Clim Chang 109:77–94. doi:10.1007/s10584-011-0151-4
Thuiller W, Lavorel S, Araújo MB, Sykes MT, Prentice IC (2005) Climate change threats to plant diversity in Europe. Proc Natl Acad Sci U S A 102:8245–8250. doi:10.1073/pnas.0409902102
Tkach N, Ree RH, Kuss P, Roser M, Hoffmann MH (2014) High mountain origin, phylogenetics, evolution, and niche conservatism of arctic lineages in the hemiparasitic genus Pedicularis (Orobanchaceae). Mol Phylogenet Evol 76:75–92. doi:10.1016/j.ympev.2014.03.004
Tremblay NO, Schoen J (1999) Molecular phylogeography of Dryas integrifolia: glacial refugia and postglacial recolonization. Mol Ecol 8:1187–1198. doi:10.1046/j.1365-294x.1999.00680.x
Waltari E, Hijmans RJ, Peterson AT, Nyári ÁS, Perkins SL (2007) Locating Pleistocene Refugia: comparing Phylogeographic and ecological niche model predictions. PLoS One 2:e563. doi:10.1371/journal.pone.0000563
Watanabe S, Shiogama H, Tatebe H, Hayashi M, Ishii M, Kimoto M (2011) MIROC-ESM 2012: model description and basic results of CMIP5-20c3m experiments. Nat Clim Chang 4:893–897. doi:10.5194/gmd-4-845-2011
Wróblewska A (2012) The role of disjunction and postglacial population expansion on phylogeographical history and genetic diversity of the circumboreal plant Chamaedaphne calyculata. Biol J Linn Soc 105:761–775. doi:10.1111/j.1095-8312.2011.01828.x
Wróblewska A (2013a) High genetic diversity within island-like peripheral populations of Pedicularis sceptrum-carolinum, a species with a northern geographic distribution. Ann Bot Fenn 50:289–299. doi:10.5735/086.050.0511
Wróblewska A (2013b) The phylogeographical and population genetic approach to the investigation of the genetic diversity patterns in self-incompatible clonal and polyploid Linnaea borealis subsp. borealis. Bot J Linn Soc 173:64–76. doi:10.1111/boj.12073
Young A, Boyle T, Brown T (1996) The population genetic consequences of habitat fragmentation for plants. Trends Ecol Evol 11:413–418. doi:10.1016/0169-5347(96)10045-8
Acknowledgments
We thank Torbjörn Alm, Ulla-Britt Andersson, Igor Artemov, Alexander Batalov, Alan Batten, Marilyn, Barker, Bruce Bennett, Evgen Berestovskij, Ragna Breines, Emilia Brzosko, Alicja Buczek, Gheorghe Coldea, Thierry Delahaye, Jennifer Doubt, Peter Efimov, Arve Elvebakk, Dorothee Ehrich, Dorothy Fabian, Jeremie Fant, Noriyuki Fujii, Bruno Haas, Edyta Jermakowicz, Natalia Koroleva, Marek Krasowski, Patrick Kuss, Martin Lechowicz, Martin Magnes, Joy Marburger, Wiaczesław Michalczuk, Anna Otręba, Carolyn Parker, Renata Piwowarczyk, Laura Poggio, Michał Ronikier, Martti Salo, Michael Schafale, Andrew Scobie, Peter Scott, Vladimir Semerikov, Przemysław Stachyra, Monika Szczecińska, Oleg Sozinov, Adrian Stoica, Kadri Tali, Izabela Tałałaj, Irina Tatarenko, Gergely Várkonyi, Andy Walker, Anna Wedel-Sala, Kristine Bakke Westtergaard, Dan Wołkowycki, Christopher C. Wilcock, and Hanna Zacharczuk for helping to collect samples, Dorothee Ehrich for statistical analysis based on the R script and Beata Ostrowiecka for technical assistance in the laboratory. We would like to thank two anonymous reviewers for their valuable suggestions and critiques of the manuscript. This research was funded by a grant from the Polish Ministry of Science and Higher Education (NN 303 366135).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Data accessibility
DNA sequences have been submitted to GenBank with no. KT182410-KT182433. AFLP data and climate details were deposited in Dryad: http://dx.doi.org/10.5061/dryad.n9h1g.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Wróblewska, A., Mirski, P. From past to future: impact of climate change on range shifts and genetic diversity patterns of circumboreal plants. Reg Environ Change 18, 409–424 (2018). https://doi.org/10.1007/s10113-017-1208-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10113-017-1208-3