Abstract
Epidemiological surveillance of Shigella spp. in Australia is conducted to inform public health response. Multi-drug resistance has recently emerged as a contributing factor to sustained local transmission of Shigella spp. All data were collected as part of routine public health surveillance, and strains were whole-genome sequenced for further molecular characterisation. 108 patients with an endemic regional Shigella flexneri strain were identified between 2016 and 2019. The S. flexneri phylogroup 3 strain endemic to northern Australia acquired a multi-drug resistance conferring blaDHA plasmid, which has an IncFII plasmid backbone with virulence and resistance elements typically found in IncR plasmids. This is the first report of multi-drug resistance in Shigella sp. in Australia that is not associated with men who have sex with men. This strain caused an outbreak of multi-drug-resistant S. flexneri in northern Australia that disproportionality affects Aboriginal and Torres Strait Islander children. Community controlled public health action is recommended.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Shigellosis in Australia is generally understood to occur as sporadic cases among returning travellers or amongst men who have sex with men (MSM) as locally acquired, sexually transmitted infections [1]. However, in northern Queensland and the Northern Territory, locally acquired strains of both Shigella sonnei and S. flexneri are persistently transmitted within other population groups [2]. These endemic strains have previously been mostly susceptible to commonly used antimicrobial agents including ampicillin, ciprofloxacin and co-trimoxazole [3].
In 2019, both Australia and Queensland had a shigellosis notification rate of 12 per 100,000 head of population, while in contrast, the Northern Territory had a notification rate of 119 per 100,000 [4], and in the northernmost health and hospital service districts of Queensland (Torres and Cape, Cairns and Hinterland, North West, and Townsville [5]), the notification rate was 24.7 per 100,000. Northern Queensland and the Northern Territory cover an area of approximately 1.9 million km2, much of which is sparsely populated and geographically remote. The population of northern Australia has a large proportion of Aboriginal and Torres Strait Islander peoples who are uniquely vulnerable to diarrhoeal disease for a variety of reasons including significant social disadvantage and inadequate housing conditions [6, 7]. Children of remote communities in this region experience a disproportionate infectious disease burden when compared with the entire Australian population [8], including for trachoma which, although is understood to be eliminated in Queensland, is still endemic in the Northern Territory and can also be managed with azithromycin [9, 10].
The emergence of MDR strains in MSM was a driver for guideline changes in 2019; subsequently, Australian therapeutic guidelines now recommend treating shigellosis with ceftriaxone in immunocompromised patients or those with severe infection, or where required for those who are high risk, whilst laboratory phenotypic AMR testing is pending [11]. Prior to this recent change, the recommended agents were co-trimoxazole, ciprofloxacin or azithromycin. Notification to CARAlert, the national alert system for critical antimicrobial resistance (AMR), is currently required if phenotypic resistance is observed to any three of the following: ampicillin/amoxicillin, ciprofloxacin/norfloxacin, co-trimoxazole, ceftriaxone/cefotaxime/ceftazidime (3GCR) or azithromycin with molecular methods for characterisation of the beta-lactamase gene/s as either AmpC-like (class C plasmid-mediated resistance) or ESBL (extended-spectrum beta-lactamase) is recommended but not required for notification. Prior to 2019, azithromycin was not one of the categories listed in the laboratory handbook [12].
Plasmids play a pivotal role in the acquisition and dissemination AMR via mobile genetic elements (MGE), with IncF plasmids being predominant types among Enterobacterales [13]. A study into the global dissemination of MDR S. flexneri among MSM found that the IncFII35 plasmid pKSR100 is associated with resistant lineages [8]. This plasmid carries mph(A) and ermB conferring resistance to azithromycin, as well as other AMR determinants. Related plasmids have subsequently been found and are mainly associated with MSM in other MDR lineages globally [14], including IncFII35 pKSR100–like plasmid described in MSM associated S. flexneri 2a in metropolitan south-east Australia [1]. The IncFII2 R100 plasmid from S. flexneri from Japan in the 1950s is considered the classical IncFII plasmid [13] and contains tet(A) and catA AMR genes.
The association between virulence and AMR genes facilitates the persistence of pathogenic bacteria leading to the development of high-risk clones [15]. The phage shock protein (PSP) system mediated by the psp operon is an integral part of the stress response, enabling bacteria to compete for survival under energy-limited conditions. The IncR pKSP30 from Klebsiella pneumoniae carries the sap operon (plays a role in resistance to cationic peptides), psp operon and 3’CS extreme of a class 1 integron consisting of qacEΔ1 conferring resistance to quaternary ammonium compounds, followed by sul1 along with other MDR determinants [16, 17]. The presence of atypical class 1 integrons is commonly reported in Shigella spp. from Asia and Africa [18,19,20].
Whole-genome sequencing (WGS) is established as the method of choice for identification and typing of Shigella spp. [21]. A species defining study has divided clonal complex 245 (CC245) S. flexneri (all S. flexneri serogroups other than O6) into seven phylogroups (PGs) by core single nucleotide polymorphism (SNP) typing analysis [22]. Genes responsible for determining S. flexneri serotypes are encoded on horizontally transmissible elements and can easily be lost or gained under selective pressure; thus, serotype results may not represent true phylogenetic relationships [23]. Queensland Health Forensic and Scientific Services Public Health Microbiology is the state reference laboratory for Shigella spp. isolated in Queensland, undertaking routine WGS surveillance of all Shigella spp. isolates received, which consists of approximately 40% of total Shigellosis notifications in the state. The remainder of notifications is PCR-only diagnoses, with no isolate available for epidemiological typing. This routine surveillance has identified the persistence of an endemic local strain of PG3 S. flexneri in northern Queensland and the Northern Territory among 108 different patients between 2016 and 2019.
Materials and methods
Data analysis
All data was collected as part of routine health surveillance activities, outbreak support and reporting between 2016 and 2019 under the Queensland Public Health Act [24], (supplementary Table 1). Rates are calculated as crude rates using population data sourced from the Australian Bureau of Statistics [25].
Laboratory phenotyping
All isolates were serotyped with agglutinating sera (Denka Seiken, Japan) and, where antimicrobial susceptibility results were available, performed with Vitek 2 GN AST card and Etests for azithromycin (bioMérieux, France) using EUCAST [26] interpretations.
Laboratory genotyping
WGS was performed using the Nextera XT library preparation kit and the Nextseq 500 platform (Illumina, CA) and reads trimmed with trimmomatic [27] and assembled with Spades [28]. Core SNP analysis was performed using snippy [29] with S. flexneri serotype 2a PG2 strain 301 chromosome NC_004337.2 as a reference. The presence of AMR determinants was determined with resfinder and point finder [30]. One strain (M2901) was selected for promethION sequencing (Oxford Nanopore Technologies, UK). Nanopore sequencing libraries were prepared using the rapid barcode sequencing kit SQK-RBK004 and sequenced on a single flow cell (R9.4.1) for 48 h. Raw Nanopore sequence data was de novo assembled using Flye with default parameters [31]. The draft genome was polished with Medaka [32], followed by polishing with both long and short reads using NextPolish v1.2.4 [33]. Annotation was performed with NCBI prokaryotic genome annotation pipeline [34].
Results
Epidemiology
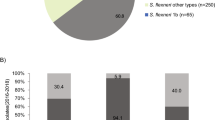
The rate of shigellosis notifications for Aboriginal and Torres Strait Island people living in north Queensland has been consistently higher than observed for non-Indigenous residents of this region. This is most evident in the three years from 2017 to 2019, with 76 notifications per 100,000 population in Aboriginal and Torres Strait Islander residents compared with 11 notifications per 100,000 in non-Indigenous residents, and even more striking for S. flexneri (Fig. 1).
Of the 108 patients with strains belonging to the endemic northern Australia PG3 S. flexneri clone, 103 (95%) resided in either the Northern Territory or one of the four northernmost health and hospital service districts of Queensland. The median age of patients was 14 years which is significantly lower than the median age of 33 years for all Shigellosis notifications for the outbreak periods 2016 to 2019 from Queensland (Kruskal-Wallis H = 42.0; P < 0.001).
Laboratory results
Although most strains in the outbreak were serotype 2b, five were var X and four were 2a; however, all belonged to the same phylogroup (PG3) and clustered together by core SNP typing analysis. Figure 2 shows a phylogenetic tree built using SNP typing with the outbreak strains, compared with a publicly available PG3 S. flexneri 2a MDR strain AUSMDU00008332 associated with MSM in south-east Australia and globally, as well as the PG1 S. flexneri 1c strain AUSMDU00008355. The pKSR100-like plasmid from strain AUSMDU00008332 does not possess drfA17, sul1 and aadA5; however, the one from AUSMDU00008355 does. Plasmid MLST [38] performed in silico via pubMLST revealed all 108 strains in the outbreak to have an IncFII27 virulence plasmid carrying the invasion plasmid antigen IpaH. In this study, the outbreak strain appears to be established and endemic in the northern Australia region since 2016 but has acquired MDR determinants by gaining an additional blaDHA carrying IncFII2 plasmid (pM2901) more recently in 2018. Prior to this, plasmid-based MGE associated with AMR has appeared in this clone; however, none has established themselves like the blaDHA plasmid. All strains were found to carry blaOXA-1, tet(B), catA1 and dfrA1, plus at least one gene conferring aminoglycoside resistance while they expressed phenotypic resistance to ampicillin and/or amoxicillin. In addition to pM2901, some strains also carried an additional IncI114 plasmid carrying blaTEM1-C and sul2. All AMR conferring genes cited in this manuscript are listed with the class of AMR they are expected to confer resistance to in Table 1.
Phylogenetic tree of core SNP typing built using snippy [29] and RAxML [35], visualised with iTOL [36], showing PG3 outbreak strains with acquired AMR genes detected, including strain AUSMDU00008332 accession NZ_LR213455.1 from the global MDR MSM outbreak of PG3 S. flexneri 2a, and AUSMDU00008355 accession NZ_LR213452.1 from PG1 S. flexneri 1c for context. Strain M2901, for which the plasmid has been characterised in Fig. 3, is marked. The year of isolation is noted.
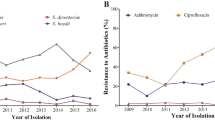
Just two strains in the outbreak with a qnrS gene expressed phenotypic quinolone resistance. Further 27 strains carried the plasmid-mediated quinolone resistance gene qnrB [39]; however, all 27 strains were classified as susceptible to quinolone antibiotics. No known quinolone resistance determining mutations were observed in any strain for gyrA or parC. Although all 108 strains had at least one trimethoprim dfrA resistance marker, with 26 of these also possessing sul1, only those strains also possessing the sul2 gene (n = 9) demonstrated co-trimoxazole resistance.
Many strains with multiple AMR markers did not meet the criteria for CARAlert notification owing to some not expressing phenotypic resistance to match their genotype. Moreover, azithromycin resistance is an additional test to those performed routinely, and if it is not tested for a Shigella sp., which is resistant to two other classes of antimicrobials, it will therefore not meet the criteria for notification even if it possesses a gene encoding for macrolide resistance. All four of the strains that showed phenotypic resistance to azithromycin when tested demonstrated the mph(A) macrolide resistance conferring gene. Although all but one outbreak strains had blaOXA-1 and were phenotypically resistant to ampicillin and/or amoxicillin, only those with an additional blaDHA/TEM/CTX-M gene showed resistance to a third-generation cephalosporin (3GCR).
Plasmid description
The pM2901 is an 81,732-bp plasmid with an average G + C content of 52.5% with transfer-encoding region. It has a backbone akin to pR100, but with an insertion of a psp operon and blaDHA, qacEΔ1, sul1 and qnrB genes similar to that of the IncR plasmid pKPS30 which carries blaOXA-1, in addition to dfrA17 like the pKSR100-like plasmids found in non-PG3 Shigella spp. in MSM in south-east Australia. Unlike the pKSR100-like plasmids, aadA5 of pM2901 was truncated with a hypothetical protein. Additionally, the tet(A)/catA locus of pR100 was entirely absent from pM2901. The pM2901 plasmid is most similar to p7102_58-6, which has been described in Salmonella Worthington in vivo acquisition from Citrobacter amalonaticus [40], and also pUB_DHA-1 from third-generation cephalosporin-resistant (3GCR) Escherichia coli in England [41]. Figure 3 shows an alignment of the pM2901 against other plasmids.
Outbreak associated plasmid pM2901 aligned using BLASTn, as implemented in Easyfig [37], against (A) pR100 IncFII2 accession AP000342 and pKSP30 IncR accession KF793937. (B) pKSR100 IncFII35 accession LN624486 and pKSR100-like IncFII35 accession NZ_LR213453.1 from strain AUSMDU00008355
Sequence data for strain M2910 is available at the National Center for Biotechnology Information (NCBI) BioProject PRJNA599490, and assembly data at accessions CP058589-CP058593.
Discussion
This is the first report of locally acquired MDR or azithromycin-resistant Shigella spp. in Australia that is not associated with MSM; this endemic strain disproportionately affects Aboriginal and Torres Strait Islander communities of remote Northern Territory and far-north Queensland.
The broad host range of the main plasmid involved in this outbreak, even though restricted to Enterobacterales [42], heightens its public health impact having previously been reported in Salmonella sp. and E. coli., blaDHA-1 has traditionally been found on narrow-host range IncFII plasmids; however, Mata et al. [43] have reported the co-localisation of qnrB with blaDHA-1 on broad host range L/M plasmids and an IncN plasmid. This is the first report of the co-localisation of these genes in an IncFII plasmid in Shigella sp. Wang et al. recently reported a blaIMP plasmid with a psp operon clustered with blaDHA and qnrB in an Enterobacter cloacae IncHI2 plasmid [44]. pK245 (NCBI accession NC_010886) from Klebsiella pneumoniae is considered the protype plasmid of IncR [45] and does not contain the psp operon. Although IncR plasmids are thought to be non-transferable and non-mobilisable due to lacking a transfer system and a relaxase [45], this demonstrates the transposition/plasmid recombination of IncR resistance and virulence elements to an IncF plasmid (containing a transfer system and relaxase), demonstrating the high plasticity of bacterial plasmids as anticipated by Compain et al. [46]. pHN7A8 from China JN232517 is another contemporary IncFII plasmid with a resistance region derived from R100 [47]. The dual presence of IncFII2 and IncR in Shigella sp. strains in this study demonstrates the potential for strains to carry multiple plasmid types with different incompatibility types, which was historically thought to not be possible; however, IncFII plasmids have been shown previously to diverge and overcome the incompatibility barrier with other plasmids [48]. This study did not examine the host microbiomes of patients to determine if the plasmid is hosted in organisms other than Shigella spp., limiting the insight into high-resolution plasmid relationships.
This outbreak consisted of multiple serotypes, which highlights a challenge for vaccine development where immunity to Shigella spp., is considered to be serotype-specific [49, 50]. There was no evidence in this outbreak of patients being re-infected with either the same or different serotypes or species of Shigella spp. The inclusion of different serotypes in the single cluster confirms phylogrouping as a more relevant method for public health surveillance and outbreak investigation than serotyping, as these cases would have been excluded from the investigation based on serotype results if WGS were not performed. We maintain the position that molecular genotyping of AMR is more suitable for public health surveillance purposes, while phenotyping is relevant for individual patient management as AMR genes may be present but not be expressed until selective pressure is induced by exposure to antimicrobial agents.
Without public health intervention, continued transmission of this now MDR strain of S. flexneri is likely to persist in this vulnerable population and has the potential to spread to other geographical regions. The emergence of azithromycin resistance markers in a population potentially treated with azithromycin for trachoma and/or other conditions supports the theory of Baker et al. [14] that selective pressure from azithromycin administration has contributed to the emergence of resistance in this organism. This theory is supported by the co-emergence of low-level azithromycin resistance in Neisseria gonorrhoeae (data not shown) and macrolide resistance genetic markers in Mycoplasma genitalium [51] that are currently occurring via different mechanisms in this same geographical area, and which is likely to also emerge in other infectious agents with continued social disadvantage. The recent inclusion of azithromycin as a separate category of AMR class contributing towards CARAlert notification is likely to facilitate more notifications than the previous criteria and therefore improve public health outcomes. This was evidenced by a 218% increase is CARAlert notifications for MDR Shigella spp. in Australia in 2019 from 2018, although this is primarily attributed to MDR outbreaks among MSM in other states [52]. Azithromycin testing is now required much more frequently than it was previously, for Shigella spp. from Australian patients in both urban and remote regions.
This report highlights the need for continuing AMR notification and surveillance which includes azithromycin for various organisms of public health significance among populations potentially subjected to community azithromycin distribution as a control measure for trachoma [53]. Addressing social inequalities is however the most fundamental aspect of reducing disease burden. Community controlled responses including health promotion initiatives to reduce the burden of diarrhoeal diseases, as well as rapid progression of healthy housing initiatives to reduce overcrowding and sanitation in remote communities are recommended. This is vital in improving housing, sanitation and socio-economic status and in continuing the important work of closing the gap in health and life expectancy between Aboriginal and Torres Strait Islander peoples and non-Indigenous Australians [54].
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information files.
References
Ingle DJ, Easton M, Valcanis M, Seemann T, Kwong JC, Stephens N, Carter GP, Gonçalves da Silva A, Adamopoulos J, Baines SL, Holt KE, Chow EPF, Fairley CK, Chen MY, Kirk M, Howden BP, Williamson DA (2019) Co-circulation of multidrug-resistant Shigella among men who have sex with men, Australia. Clin Infect Dis 69(9):1535–1544. https://doi.org/10.1093/cid/ciz005
Draper A, Markey P (2017) Shigella flexneri 2b in the northern territory in 2017. The Northern Territory Disease Control Bulletin 24(4):1–6
Malau E, Ford R, Valcanis M, Jennison AV, Mosse J, Bean D, Yoannes M, Pomat W, Horwood PF, Greenhill AR (2018) Antimicrobial sensitivity trends and virulence genes in Shigella spp. from the Oceania region. Infect Genet Evol 64:52–56. https://doi.org/10.1016/j.meegid.2018.06.015
National Notifiable Diseases Surveillance System (2019) Australian Government Department of Health. http://www9.health.gov.au/cda/source/cda-index.cfm. Accessed 16 July 2019
Hospital and Health Service maps (2020) Queensland Health. https://www.health.qld.gov.au/maps. Accessed 5 July 2020
Falster K, Banks E, Lujic S, Falster M, Lynch J, Zwi K, Eades S, Leyland AH, Jorm L (2016) Inequalities in pediatric avoidable hospitalizations between Aboriginal and non-Aboriginal children in Australia: a population data linkage study. BMC Pediatr 16(1):169. https://doi.org/10.1186/s12887-016-0706-7
Foster T, Hall NL (2019) Housing conditions and health in Indigenous Australian communities: current status and recent trends. Int J Environ Health Res:1–19. https://doi.org/10.1080/09603123.2019.1657074
Clucas DB, Carville KS, Connors C, Currie BJ, Carapetis JR, Andrews RM (2008) Disease burden and health-care clinic attendances for young children in remote aboriginal communities of northern Australia. Bull World Health Organ 86:275–281. https://doi.org/10.2471/BLT.07.043034
Australian Trachoma Surveillance Report 2017 (2018) Kirby Institute UNSW, Sydney
CDNA National Guidelines for the Public Health Management of Trachoma (2014) Communicable diseases network Australia
eTG complete [digital] (2019) https://www.tg.org.au. Accessed 2019 Dec
CARAlert Laboratory Handbook (2019) Australia Commission of Safety and Quality in Health Care, Sydney
Partridge SR, Kwong SM, Firth N, Jensen SO (2018) Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev 31(4):e00088–e00017. https://doi.org/10.1128/CMR.00088-17
Baker KS, Dallman TJ, Ashton PM, Day M, Hughes G, Crook PD, Gilbart VL, Zittermann S, Allen VG, Howden BP (2015) Intercontinental dissemination of azithromycin-resistant shigellosis through sexual transmission: a cross-sectional study. Lancet Infect Dis 15(8):913–921. https://doi.org/10.1016/S1473-3099(15)00002-X
Beceiro A, Tomás M, Bou G (2013) Antimicrobial resistance and virulence: a successful or deleterious association in the bacterial world? Clin Microbiol Rev 26(2):185–230. https://doi.org/10.1128/CMR.00059-12
Darwin AJ (2005) The phage-shock-protein response. Mol Microbiol 57(3):621–628. https://doi.org/10.1111/j.1365-2958.2005.04694.x
Paulsen I, Littlejohn T, Rådström P, Sundström L, Sköld O, Swedberg G, Skurray R (1993) The 3’ conserved segment of integrons contains a gene associated with multidrug resistance to antiseptics and disinfectants. Antimicrob Agents Chemother 37(4):761–768. https://doi.org/10.1128/AAC.37.4.761
Kang H, Wang L, Li Y, Lu Y, Fan W, Bi R, Qian H, Gu B (2019) Dissemination of multidrug-resistant Shigella flexneri and Shigella sonnei with class 1, class 2, and atypical class 1 integrons in China. Microb Drug Resist 25(10):1465–1474. https://doi.org/10.1089/mdr.2018.0229
Xu Y, Zhuang L, Kang H, Ma P, Xu T, Pan S, Gu B (2016) Prevalence, resistance patterns, and characterization of integrons of Shigella flexneri isolated from Jiangsu Province in China, 2001–2011. Eur J Clin Microbiol Infect Dis 35(8):1347–1353. https://doi.org/10.1007/s10096-016-2671-3
Schaumburg F, Alabi AS, Kaba H, Lell B, Becker K, Grobusch MP, Kremsner PG, Mellmann A (2015) Molecular characterization of Shigella spp. from patients in Gabon 2011–2013. Trans R Soc Trop Med Hyg 109(4):275–279. https://doi.org/10.1093/trstmh/tru175
Chattaway MA, Schaefer U, Tewolde R, Dallman TJ, Jenkins C (2017) Identification of Escherichia coli and Shigella species from whole genome sequences. J Clin Microbiol 55(2):616–623. https://doi.org/10.1128/jcm.01790-16
Connor TR, Barker CR, Baker KS, Weill F-X, Talukder KA, Smith AM, Baker S, Gouali M, Thanh DP, Azmi IJ (2015) Species-wide whole genome sequencing reveals historical global spread and recent local persistence in Shigella flexneri. Elife 4:e07335. https://doi.org/10.7554/eLife.07335
Allison GE, Verma NK (2000) Serotype-converting bacteriophages and O-antigen modification in Shigella flexneri. Trends Microbiol 8(1):17–23. https://doi.org/10.1016/S0966-842X(99)01646-7
Public Health Act (2005) Queensland Government
Regional Population by Age and Sex, Australia 2017 (2018) 3235.0 edn. Australian Bureau of Statistics
Breakpoint tables for interpretation of MICs and zone diameters (2019) The European Committee on Antimicrobial Susceptibility Testing
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19(5):455–477. https://doi.org/10.1089/cmb.2012.0021
Seeman T (2015) Snippy: rapid haploid variant calling and core SNP phylogeny, 4th edn. https://github.com/tseemann/snippy Accessed 5 July 2020
Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV (2012) Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67(11):2640–2644. https://doi.org/10.1093/jac/dks261
Kolmogorov M, Yuan J, Lin Y, Pevzner PA (2019) Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol 37(5):540–546. https://doi.org/10.1101/247148
Medaka (2018) 1.0.3 edn. Oxford Nanopore Technologies
Hu J, Fan J, Sun Z, Liu S (2020) NextPolish: a fast and efficient genome polishing tool for long-read assembly. Bioinformatics 36(7):2253–2255. https://doi.org/10.1093/bioinformatics/btz891
Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J (2016) NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44(14):6614–6624. https://doi.org/10.1093/nar/gkw569
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Letunic I, Bork P (2016) Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44(W1):W242–W245. https://doi.org/10.1093/nar/gkw290
Sullivan MJ, Petty NK, Beatson SA (2011) Easyfig: a genome comparison visualizer. Bioinformatics 27(7):1009–1010. https://doi.org/10.1093/bioinformatics/btr039
Villa L, García-Fernández A, Fortini D, Carattoli A (2010) Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother 65(12):2518–2529. https://doi.org/10.1093/jac/dkq347
Jacoby GA, Walsh KE, Mills DM, Walker VJ, Oh H, Robicsek A, Hooper DC (2006) qnrB, another plasmid-mediated gene for quinolone resistance. Antimicrob Agents Chemother 50(4):1178–1182. https://doi.org/10.1128/AAC.50.4.1178-1182.2006
Clément M, Keller PM, Bernasconi OJ, Stirnimann G, Frey PM, Bloemberg GV, Sendi P, Endimiani A (2019) First clinical case of in vivo acquisition of DHA-1 plasmid-mediated AmpC in a Salmonella enterica subsp. enterica isolate. Antimicrob Agents Chemother 63(10):e00992–e00919. https://doi.org/10.1128/AAC.00992-19
Findlay J, Gould VC, North P, Bowker KE, Williams MO, MacGowan AP, Avison MB (2019) Characterization of cefotaxime-resistant urinary Escherichia coli from primary care in South-West England 2017–18. J Antimicrob Chemother 75(1):65–71. https://doi.org/10.1093/jac/dkz397
Carattoli A (2009) Resistance plasmid families in Enterobacteriaceae. Antimicrob Agents Chemother 53(6):2227–2238. https://doi.org/10.1128/AAC.01707-08
Mata C, Miró E, Toleman M, Rivera M, Walsh TR, Navarro F (2011) Association of blaDHA-1 and qnrB genes carried by broad-host-range plasmids among isolates of Enterobacteriaceae at a Spanish hospital. Clin Microbiol Infect 17(10):1514–1517. https://doi.org/10.1111/j.1469-0691.2011.03539.x
Wang S, Zhou K, Xiao S, Xie L, Gu F, Li X, Ni Y, Sun J, Han L (2019) A multidrug resistance plasmid pIMP26, carrying blaIMP-26, fosA5, blaDHA-1, and qnrB4 in Enterobacter cloacae. Sci Rep 9(1):1–7. https://doi.org/10.1038/s41598-019-46777-6
Alvarado A, Garcillán-Barcia MP, de la Cruz F (2012) A degenerate primer MOB typing (DPMT) method to classify gamma-proteobacterial plasmids in clinical and environmental settings. PLoS One 7(7):e40438. https://doi.org/10.1371/journal.pone.0040438
Compain F, Frangeul L, Drieux L, Verdet C, Brisse S, Arlet G, Decré D (2014) Complete nucleotide sequence of two multidrug-resistant IncR plasmids from Klebsiella pneumoniae. Antimicrob Agents Chemother 58(7):4207–4210. https://doi.org/10.1128/AAC.02773-13
He L, Partridge SR, Yang X, Hou J, Deng Y, Yao Q, Zeng Z, Chen Z, Liu J-H (2012) Complete nucleotide sequence of pHN7A8, an F33: A-: B-type epidemic plasmid carrying blaCTX-M-65, fosA3 and rmtB from China. J Antimicrob Chemother 68(1):46–50. https://doi.org/10.1093/jac/dks369
Osborn AM, da Silva Tatley FM, Steyn LM, Pickup RW, Saunders JR (2000) Mosaic plasmids and mosaic replicons: evolutionary lessons from the analysis of genetic diversity in IncFII-related replicons. Microbiology 146(9):2267–2275. https://doi.org/10.1099/00221287-146-9-2267
Cohen D, Green M, Block C, Slepon R, Ofek I (1991) Prospective study of the association between serum antibodies to lipopolysaccharide O antigen and the attack rate of shigellosis. J Clin Microbiol 29(2):386–389
Levine MM, Kotloff KL, Barry EM, Pasetti MF, Sztein MB (2007) Clinical trials of Shigella vaccines: two steps forward and one step back on a long, hard road. Nat Rev Microbiol 5(7):540–553. https://doi.org/10.1038/nrmicro1662
Sweeney E, Trembizki E, Bletchly C, Bradshaw C, Menon A, Francis F, Langton-Lockton J, Nimmo G, Whiley D (2019) Levels of Mycoplasma genitalium antimicrobial resistance differ by both region and gender in the state of Queensland, Australia: implications for treatment guidelines. J Clin Microbiol 57(3):e01555–e01518. https://doi.org/10.1128/JCM.01555-18
CARAlert annual report: 2019 (2020) Australian Commission on Safety and Quality in Health Care, Sydney
O’Brien KS, Emerson P, Hooper PJ, Reingold AL, Dennis EG, Keenan JD, Lietman TM, Oldenburg CE (2019) Antimicrobial resistance following mass azithromycin distribution for trachoma: a systematic review. Lancet Infect Dis 19(1):e14–e25. https://doi.org/10.1016/s1473-3099(18)30444-4
McDonald EL (2011) Closing the gap and indigenous housing. Med J Aust 195(11):652–653. https://doi.org/10.5694/mja11.10376
Jolley KA, Bray JE, Maiden MCJ (2018) Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res 3:124. https://doi.org/10.12688/wellcomeopenres.14826.1
Acknowledgements
This publication made use of the plasmid MLST website (https://pubmlst.org/plasmid/) sited at the University of Oxford [55]. The development of this site has been funded by the Wellcome Trust.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
This is an observational study. The Queensland Health Forensic and Scientific Services Human Ethics Committee has confirmed that a full ethics review is not required, and ethics clearance has been granted.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Code availability
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary Table 1.
Line list of outbreak strains with AMR genes and phenotypic profiles. (XLSX 38 kb).
Rights and permissions
About this article
Cite this article
Guglielmino, C.J.D., Kakkanat, A., Forde, B.M. et al. Outbreak of multi-drug-resistant (MDR) Shigella flexneri in northern Australia due to an endemic regional clone acquiring an IncFII plasmid. Eur J Clin Microbiol Infect Dis 40, 279–286 (2021). https://doi.org/10.1007/s10096-020-04029-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-020-04029-w