Abstract
Syndromic panel-based molecular testing has been suggested to improve and accelerate microbiological diagnosis. We aimed to analyze workflow improvements when using the multiplex Seegene Allplex™ GI-Bacteria(I) assay as a first-line assay for bacterial diarrhea. Technical assay evaluation was done using spiked stool samples and stored patient samples. After implementation of the assay in the routine clinical workflow, an analysis of 5032 clinical samples analyzed by the Seegene assay and 4173 control samples examined by culture in a similar time period 1 year earlier was performed. Sensitivity of the assay was shown to be between 0.4 and 95.9 genome equivalents/PCR. For 159 positive patient samples with a composite reference of culture and/or a molecular assay, the sensitivity of the assay was 100% for Campylobacter, 92% for Salmonella, 89% for Aeromonas, and 83% for Shigella. Sensitivity for C. difficile toxin B detection was 93.9%. The comparison of clinical samples obtained in two 8-month periods showed increased detection rates for Aeromonas (2.90%vs. 0.34%), Campylobacter spp. (2.25% vs. 1.34%), Shigella spp. (0.42% vs. 0.05%) whereas detection of Salmonella was slightly decreased (0.46% vs. 0.67%) when using the Seegene assay. An analysis of the time-to-result showed that the median dropped from 52.7 to 26.4 h when using the molecular panel testing. The Seegene Allplex™ GI-Bacteria(I) assay allows accelerated, reliable detection of major gastrointestinal bacteria roughly within 1 day. Workload is reduced, specifically in a low-prevalence setting.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Syndromic testing for infectious diseases, based on multiplexed molecular assays, has recently been introduced into clinical microbiology [1]. Amongst other diseases, gastrointestinal infections are addressed by a number of commercialized assays, some of them with an approval by the US food and drug administration [2]. Those assays detect multiple pathogens in parallel that can cause diarrhea. Routinely, diarrheal pathogens are identified by a combination of microscopy, antigen testing, culture, and singleplex PCR. The main disadvantage of the conventional approach is time-to-result, which for culture can take days, even for negative findings. Meanwhile multiple singleplex PCRs are available, but as clinical signs are often unspecific, ordering of multiple tests becomes uneconomic. Thus, syndromic, multiplex-based assays are promising to have clinical impact [3]. Advantages that are associated with syndromic, molecular testing include a rapid turnaround time that can affect clinical decision strategies including hospital admission, isolation, and infection control measures as well as sensitivities that often are superior to testing by culture [1]. Moreover, various pathogens can be detected in parallel, if clinical symptoms are unspecific. On the other hand, multiplex panels are expensive and clinical implementation strategies still have to be developed [4]. Panel compositions vary, but are mostly fixed; thus, laboratories will also have to cope with selecting panels that appropriately cover the local microorganisms. For diarrheal disease which is mostly self-limiting, routine testing for pathogens is not recommended but may be used in case of comorbidities, immunosuppression, bloody diarrhea, severe illness, decision of hospital admission, patients from community facilities, travelers, or patients with prolonged symptoms > 7 days [5, 6]. If available, an FDA-approved culture-independent method is recommended as an adjunct to conventional methods [5].
The Seegene Allplex real-time PCR assays allow simultaneous detection of up to seven pathogens within one reaction tube using a specific detection algorithm. Based on multiple quantification by real-time PCR and a specific interpretation software, for gastrointestinal infections, four panels, altogether comprising 25 pathogens, that can either be run in a combined or selected manner, are offered. Only few data are available for the Seegene Allplex Gastrointestinal assay so far [7, 8]: In comparison with Luminex xTAG GPP and BD MAX Enteric assays, the overall positive percentage agreements of Seegene, Luminex, and BD MAX were found to be 94%, 92%, and 78% [7]. No data on implementation of this assay in a routine workflow is available by now. We therefore report here on the implementation of the bacteria (I) panel, covering Campylobacter spp., Clostridioides difficile toxin B gene, Salmonella spp., Shigella spp./EIEC, Vibrio spp., Yersina enterocolitica, and Aeromonas spp., instead of culture as a primary test for major bacterial diarrhea pathogens. Change of the diagnostic procedure was necessary to facilitate workflow, increase productivity, and reduce turnaround times for results, aims that have all been reported to be achievable by use of multiplex panel testing. A decision for the Seegene Allplex GB(I) assay was done based on cost estimates as the targets of the assay can be adopted to the local needs based on the use of selected panel tubes and as costs are lower than for fully automated, closed, but fixed systems.
Materials and methods
Multiplex PCR for syndromic panel diagnostic
Stool samples were analyzed using the Seegene Allplex™ GI-Bacteria (I) assay (Seegene, Seoul, South Korea) in combination with automated DNA extraction and PCR setup (Nimbus system) according to the manufacturer’s instructions. In brief, 150–200 μl fluid stool (equaling to 100–200 mg) were transferred from the stool container in 1 ml ASL buffer in a 2-ml tube (Qiagen, Hilden, Germany) using a flocked swab (PurFlock Ultra, Check Diagnostics, Germany) (Using a calibrated loop did not transfer enough material). The sample was vortexed, incubated for 10 min at room temperature, and then centrifuged for 2 min at 14,000 rpm. The 2-ml tube was directly used for DNA extraction. Alternatively, 800 μl supernatant was transferred into a new tube, if the ASL sample was very inhomogeneous after centrifugation. DNA extraction and PCR setup were done using STARMag Universal Cartridge kit (Seegene, Duesseldorf, Germany) in the Microlab Nimbus (Seegene) automated liquid handling workstation. The positive control was added after the automated PCR setup was manually done. The plate was removed from the Nimbus system, sealed with caps, and briefly centrifuged before analysis in a CFX96 cycler (Bio Rad, Germany). Results of the analysis were done using the Seegene Viewer software. Positive detection of Salmonella spp., Yersina enterocolitica, Campylobacter spp., Shigella spp./EIEC, and Vibrio spp. was followed by an attempt to cultivate the respective pathogen from the original stool sample (“reflective culture”). Inhibited samples were diluted 1:3 in PBS before adding into ASL buffer and then repeated once.
Detection by culture
Stool samples were analyzed according to routine procedures in the Institute of Medical Microbiology and Hygiene, Heidelberg, holding an accreditation according to DIN EN ISO 15189. In brief, stool samples were analyzed using blood (Becton Dickinson, Heidelberg, Germany), CIN, XLD agar, Campy (all bioMérieux, Marcy l’Étoile, France) selective agar, and a selenite broth (Becton Dickinson, Heidelberg, Germany), and were incubated for 24 and 48 h at 36 °C. Identification of suspicious colonies was done by MALDI-TOF (Bruker Daltonik, Bremen, Germany) using direct smear on target procedure. For further specification and confirmation, agglutination tests were performed for Salmonella spp., Shigella spp., and Yersinia spp. with specific antisera (SIFIN diagnostics, Berlin, Germany).
C. difficile detection
C. difficile toxin B detection (from here on referred to as C. difficile detection) in routine clinical samples was done directly from stool samples using the molecular BD MAX C.diff assay according to the manufacturer’s instructions.
Technical validation of the multiplex PCR assay’s performance
For determination of the limit of detection, 400 μl of a homogenous stool solution, tested negative for the respective pathogens, was spiked with 2 × 50μl of a 0.5 McF suspension of two pathogens each: C. jejuni (DSM4688), S. cholerasuis (ATCC554), Y. enterocolitica (ATCC9610), S. flexneri (ATCC29903), C. difficile (DSM27544), Aeromonas hydrophila (DSM30187), Vibrio cholerae (DSM100200). Thereafter, a 1:10 dilution series was produced (1 E7/ml to 1 E0/ml) and tested by the multiplex PCR as well as colony counting (for exact CFU determination) on the respective agar media. The test was repeated once with a 1:10 dilution series and then five times with a 1:3 dilution around the limit of detection. Moreover, 159 clinical stool samples (samples from the routine diagnostics in Heidelberg and samples from Lucerne) that had been analyzed before and that had been stored at − 80 °C for up 2 years were tested. For these samples, a culture result and/or a molecular result (BD MAX Enteric Panel, Biofire Filmarray, BD MAX C.diff) were available. In case of discrepant results, the samples were reanalyzed once by the same method. A composite of culture and/or molecular result was used as reference for the evaluation of the Seegene multiplex assay.
Multiplex PCR implementation in routine diagnostics and clinical performance validation
The Seegene multiplex PCR was implemented as standard diagnostic procedure for requests of bacteria-induced diarrhea in 2017. Stool samples are analyzed once daily from Monday to Friday. At the weekends, stool samples are diluted in ASL buffer but stored until Monday (increased storage time was evaluated not to affect assay performance). At weekends, C. difficile detection was done using the BD MAX C.diff assay, whereas at weekdays, the C. difficile result as available from the Seegene multiplex PCR assay is reported. We did a comparison of the detection rates of the respective pathogens in a time period from November 6, 2017 to July 15, 2018, when using the Seegene multiplex PCR as the primary assay, followed by culture in case of positive detections, to the same time period 1 year before (November 6, 2017–July 15, 2017) when detection was done by culture only. Time-to-result was analyzed from laboratory information system (Swisslab, Nexus AG, Berlin) by using the entries “sample received” and “validation of final report”.
Results
Technical performance of the Seegene Allplex GI-B(I) multiplex PCR
We first determined the detection limit by spiking negative stool samples with a defined concentration of the included bacteria. It could be shown that the limit of detection (90% detection rate) was between 0.4 and 95.9 genome equivalents/PCR which is in the range reported by the manufacturer. Due to the dilution of the sample in the process of detection (1:10 for preparation in ASL buffer, 5 μl/100 μl of the nucleic acid eluate within the PCR reaction), the assay detected in detail: 1.4 E5 CFU/ml for Campylobacter spp., 3.7 E4 CFU/ml Salmonella spp., 9.5 E2 CFU/ml for Shigella spp., 1.4 E4 CFU/ml for Yersina enterocolitica, and 5.7 E3 CFU/ml for Aeromonas spp. Next, we tested the assay with N = 159 clinical samples for which a positive result of the included bacteria had been obtained earlier. A compound reference of culture and/or positivity by a molecular assay was used as standard. The sensitivity of the assay was 100% for Campylobacter spp., 92% for Salmonella spp., and 89% for Aeromonas spp. For Yersinia enterocolitica, the sample numbers were small: 3 samples, positive by culture were detected by the multiplex PCR, whereas 4 samples for which only a molecular assay was positive were negative with the Seegene multiplex PCR. For Shigella spp., detection sensitivity was 100% when compared with culture but 83% when another molecular test was added. Sensitivity for C. difficile detection was evaluated by a comparison with the BD MAX C.diff kit, a singleplex PCR, and was 93.9%.
Routine results with a multiplex PCR workflow
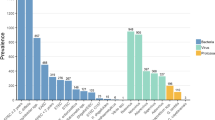
The Seegene multiplex PCR was introduced into the routine workflow as the standard diagnostic procedure replacing primary bacterial culture. Stool samples were analyzed as a batch (20–50 samples) once daily at weekdays; no diagnostic was offered at weekends. C. difficile detection was done using the Seegene multiplex PCR at weekdays, but by singleplex BD MAX PCR at weekends. The workflow allowed a considerable decrease in hands-on times as preparation and setup of the test was largely automated (estimate 3 h/day for panel diagnostic vs. 1 FTE for culture handling). Within a low-prevalence setting, the workflow resulted in only few cultures that were needed to be set up as confirmation for samples positive by the molecular assay. To evaluate whether molecular detection also increased detection rates of classical bacterial diarrhea pathogens, we compared results from an 8-month time period using the molecular assay with a similar period in the year before using culture. Of note, within this time period, the reported German-wide detection rates for the included pathogens (data from the national health authorities) were comparable. With the introduction, we observed increased detection rates for Aeromonas spp. (8.5-fold increase, 2.90% vs. 0.34%), Campylobacter spp. (1.7-fold increase, 2.25% vs. 1.34%), and Shigella spp. (8.4-fold increase, 0.42% vs. 0.05%) whereas detection of Salmonella was slightly decreased (0.46% vs. 0.67%) (Fig. 1, Table 1). C. difficile toxin B detection was 9.38% vs 10.99% by the singleplex BD MAX PCR. Upon reflex testing by culture, 56% and 61% of the positive results for Campylobacter spp. and Salmonella spp. could be confirmed (Table 1). For samples with a positive result for Shigella spp./EIEC, culture for Shigella spp. was only positive in 14% of the samples that gave a positive molecular signal. The Seegene assay uses a shared target gene for Shigella spp. and EIEC, but only for Shigella a specific culture method exists. This might contribute to the observed discrepancies. Of note, all of the tested patients had clinical symptoms of diarrhea. All positive results for Y. enterocolitica were confirmed by culture. The 7 samples positive for Vibrio were from two travelers with culture-confirmed V. cholera infection.
Performance of the Seegene Allplex™ GI-B(I) multiplex assay in a routine setting based on a before-after comparison approach. Clinical samples obtained by multiplex PCR analysis during November 2017–June 2018 (N = 5032) were compared with culture results from the corresponding previous time period November 2016–June 2017 (N = 4173). C. difficile toxin B detection was compared with a singleplex PCR
Reduced time-to-result with a multiplex PCR diagnostic algorithm
With the newly implemented molecular diagnostic algorithm, time to final result decreased from 52.7 h (median) by culture to 26.4 h by multiplex PCR (including secondary culture if necessary) (Fig. 2). The differences were significant (Kruskal-Wallis test p < 0.0001). Thus, results were available roughly 1 day earlier. Results of the PCR were obtained in 23.9% of the samples even at the same day (< 14 h) and for 74.0% of the samples within the next day (< 38 h). Of note, in the current workflow, no diagnostic by multiplex PCR was done at weekends.
Time to final result (including reflex testing) for the data of Fig. 1. Multiplex PCR, Seegene Allplex™ GI-B(I) multiplex assay, vs. culture
Discussion
We report on the implementation of the molecular Seegene Allplex™ GI-Bacteria(I) assay for primary detection of bacterial diarrheal pathogens within a low-incidence setting. The assay reliably detected major gastrointestinal bacteria including Campylobacter, Salmonella, Shigella/EIEC, and Yersinia which are to be notified to German health authorities. By performing a before-after comparison, we observed that clinical detection rates for Campylobacter, Shigella, and Aeromonas increased after introduction of the molecular panel workflow. Although a direct side-by-side comparison could not be done, the use of an equivalent time period and the data from the German health reporting system showing comparable German-wide epidemiology for the chosen periods justify such kind of comparison. Findings are in line with reports from other multiplex PCRs that show increased sensitivity for detection of gastrointestinal pathogens by molecular methods as compared with culture [9]. For the Filmarray GI Panel, a multicenter study showed detection of at least one pathogen in 54.2% of the samples vs. 18.1% with conventional techniques [10]. For the Luminex Gastrointestinal Pathogen Panel, 22.1% vs. 12% was reported [2], and other studies confirmed these differences [11, 12]. In contrast, detection of Salmonella by molecular panel was slightly less sensitive, and differences probably would have been even bigger if an enrichment broth was used. On the other hand, the Seegene assay also identified samples positive for Salmonella, for which culture was negative, indicating that both methods may find additional positive samples. Indeed, another study using the Luminex GPP assay also showed a positive percent agreement between culture and multiplex PCR for Salmonella of only 78.2% [13], with additional positive samples by both methods. Shortcomings in accuracy of molecular detection for Salmonella and Yersinia have also been reported by others [12, 14, 15]. In a comparative study with Biofire-, Luminex-, and Verigene-panels, two of the three assays had a sensitivity < 85% for detection of Salmonella whereas the other bacterial pathogens were detected very well [16]. Caution might be indicated for the use of multiplex PCR testing when specifically Salmonella shall be detected as for example in food industry workers.
Upon positive molecular detection we tried to cultivate the respective bacteria, a strategy of reflective culture. Thus, only a very limited number of samples had to undergo the time-consuming and laborious process of cultivating. For Salmonella and Campylobacter, this was only successful in 56% and 61% of the samples. As the patients suffered from diarrhea, a correct molecular diagnosis was assumed, but formally, it cannot be excluded that false-positive signals were included. Thus, it remains important that interpretation of results of multiplex panels is always done considering the patient’s symptoms, history, and risk profile [17]. Probably, not a single “gold standard” for diagnostics exists.
The workload of the molecular Seegene Allplex GI-Bacteria (I) assay was reduced as compared with the culture process which has also been reported in other studies (reviewed in [1]). One of the most important findings was the significantly reduced time-to-result with the implementation of a molecular multiplex assay. We observed a reduction of more than 1 day, and in nearly a quarter of all samples, a same-day result was achieved. Of note, various molecular panels offer a rapid detection, yet time-to-result also depends on the strategy of implementation. Here, in a routine setting, a significant reduction could be obtained. Similar savings were observed for the Biofire FilmArray with a reduction from 47 to 18 h [18]. For the Luminex assay, implemented in a routine setting, turnaround times were reduced from 66.5 to 41.8 h in one study [2], where it was noted that conventional testing for C. difficile was faster (17.3 h). In our implementation strategy, C. difficile result was obtained at latest the next day after sample delivery. Yet, as we did not offer multiplex testing at weekends, we still had to run a singleplex PCR for those indications.
As sensitivity of molecular detection is high, within a low-prevalence setting, the negative predictive value will be high, thus allowing a more rapid decision on the necessities of isolation and hygiene measurements. Based on the specific implementation, the time-to-result might be decreased further, but even with a batched protocol once a day, we had a considerable reduction. This is in line with reports for other diarrheal panels.
Implementation of a molecular assay has also to be considered under economical viewpoints. In our specific case, the change resulted in only moderately increased direct laboratory costs, because the previous procedure already involved culture plus a singleplex PCR for C. difficile, with the latter being responsible for the majority of the costs. A cost estimate for consumables was 75 K € in the 8-month period 2016–2017 (11 K € and 64 K € for culture and C. difficile detection with 4173 and 4141 samples, respectively, many of them with both requests) and 86 K € for molecular detection (71 K € and 14 K € for 5032 samples by multiplex PCR and 938 samples with singleplex C. difficile PCR). Of note, this estimate does neither consider any laboratory savings by the reduced workload nor any clinical savings. It was cost-effective, because a PCR for C. difficile could be replaced in parallel. Of course, additional economical beneficial effects come from increased diagnosis of defined pathogens and savings in isolation procedures that have not been calculated in this study. In a cost-benefit analysis for Luminex GPP testing against conventional assays, laboratory costs for 800 patients in an 8-month period increased by £22,283 but resulted in savings of £66,765, mostly due to reduction in isolation time [4]. The authors concluded that specifically, a rapid negative result could reduce cost by decreasing isolation times. Studies with gastrointestinal panels were shown to improve patient care by rapid identification of pathogens, reduction of numbers of additional test, less endoscopy and abdominal radiology, less prescription of antibiotics, and earlier release from hospital [3, 19].
Economical implementation of syndromic panel testing might also involve stratification for the use of such assay: It was shown that panel testing in patients developing diarrhea more than 3 days after hospital admission has only low yield and thus could be excluded in a decision algorithm [20].
Taken together, the Seegene Allplex™ GI-Bacteria(I) assay allowed an accelerated, reliable detection of major gastrointestinal bacteria, a reduction in the workload, and a significant decrease in the time-to-result. It may be implemented in a routine workflow as primary assay for the detection of bacteria-induced diarrhea and has advantages in a setting with high negative detection rate.
References
Ramanan P, Bryson AL, Binnicker MJ, Pritt BS, Patel R (2018) Syndromic panel-based testing in clinical microbiology. Clin Microbiol Rev 31(1):e00024
Halligan E, Edgeworth J, Bisnauthsing K, Bible J, Cliff P, Aarons E, Klein J, Patel A, Goldenberg S (2014) Multiplex molecular testing for management of infectious gastroenteritis in a hospital setting: a comparative diagnostic and clinical utility study. Clin Microbiol Infect 20(8):O460–O467
Beal SG, Tremblay EE, Toffel S, Velez L, Rand KH (2018) A gastrointestinal PCR panel improves clinical management and lowers health care costs. J Clin Microbiol 56(1):e01457
Goldenberg SD, Bacelar M, Brazier P, Bisnauthsing K, Edgeworth JD (2015) A cost benefit analysis of the Luminex xTAG gastrointestinal pathogen panel for detection of infectious gastroenteritis in hospitalised patients. J Infect 70(5):504–511
Riddle MS, DuPont HL, Connor BA (2016) ACG clinical guideline: diagnosis, treatment, and prevention of acute diarrheal infections in adults. Am J Gastroenterol 111(5):602–622
Hagel S, Epple HJ, Feurle GE, Kern WV, Lynen Jansen P, Malfertheiner P, Marth T, Meyer E, Mielke M, Moos V, von Muller L, Nattermann J, Nothacker M, Pox C, Reisinger E, Salzberger B, Salzer HJ, Weber M, Weinke T, Suerbaum S, Lohse AW, Stallmach A (2015) S2k-guideline gastrointestinal infectious diseases and Whipple’s disease. Z Gastroenterol 53(5):418–459
Yoo J, Park J, Lee HK, Yu JK, Lee GD, Park KG, Oak HC, Park YJ (2019) Comparative evaluation of Seegene Allplex gastrointestinal, Luminex xTAG gastrointestinal pathogen panel, and BD MAX enteric assays for detection of gastrointestinal pathogens in clinical stool specimens. Arch Pathol Lab Med 143(8):999–1005
Angeles Orellana Miguel M, Martín-Díaz A, Perez de Ayala Balzola A, Chaves F (2018) Performance of the multiplex-PCR assay Allplex-GI in the diagnosis of gastrointestinal infections (GI). 28th ECCMID, Madrid, p P0429
Freeman K, Mistry H, Tsertsvadze A, Royle P, McCarthy N, Taylor-Phillips S, Manuel R, Mason J (2017) Multiplex tests to identify gastrointestinal bacteria, viruses and parasites in people with suspected infectious gastroenteritis: a systematic review and economic analysis. Health Technol Assess 21(23):1–188
Spina A, Kerr KG, Cormican M, Barbut F, Eigentler A, Zerva L, Tassios P, Popescu GA, Rafila A, Eerola E, Batista J, Maass M, Aschbacher R, Olsen KEP, Allerberger F (2015) Spectrum of enteropathogens detected by the FilmArray GI panel in a multicentre study of community-acquired gastroenteritis. Clin Microbiol Infect 21(8):719–728
Buss SN, Leber A, Chapin K, Fey PD, Bankowski MJ, Jones MK, Rogatcheva M, Kanack KJ, Bourzac KM (2015) Multicenter evaluation of the BioFire FilmArray gastrointestinal panel for etiologic diagnosis of infectious gastroenteritis. J Clin Microbiol 53(3):915–925
Khare R, Espy MJ, Cebelinski E, Boxrud D, Sloan LM, Cunningham SA, Pritt BS, Patel R, Binnicker MJ (2014) Comparative evaluation of two commercial multiplex panels for detection of gastrointestinal pathogens by use of clinical stool specimens. J Clin Microbiol 52(10):3667–3673
Kellner T, Parsons B, Chui L, Berenger BM, Xie J, Burnham CA, Tarr PI, Lee BE, Nettel-Aguirre A, Szelewicki J, Vanderkooi OG, Pang XL, Zelyas N, Freedman SB (2019) Comparative evaluation of enteric bacterial culture and a molecular multiplex syndromic panel in children with acute gastroenteritis. J Clin Microbiol 57(6):e00205
Wessels E, Rusman LG, van Bussel MJAWM, Claas ECJ (2014) Added value of multiplex Luminex gastrointestinal pathogen panel (xTAG® GPP) testing in the diagnosis of infectious gastroenteritis. Clin Microbiol Infect 20(3):O182–O187
Pankhurst L, Macfarlane-Smith L, Buchanan J, Anson L, Davies K, O'Connor L, Ashwin H, Pike G, Dingle KE, Peto TE, Wordsworth S, Walker AS, Wilcox MH, Crook DW (2014) Can rapid integrated polymerase chain reaction-based diagnostics for gastrointestinal pathogens improve routine hospital infection control practice? A diagnostic study. Health Technol Assess 18(53):1–167
Huang RSP, Johnson CL, Pritchard L, Hepler R, Ton TT, Dunn JJ (2016) Performance of the Verigene® enteric pathogens test, Biofire FilmArray™ gastrointestinal panel and Luminex xTAG® gastrointestinal pathogen panel for detection of common enteric pathogens. Diagn Microbiol Infect Dis 86(4):336–339
Zautner AE, Gross U, Emele MF, Hagen RM, Frickmann H (2017) More pathogenicity or just more pathogens?-on the interpretation problem of multiple pathogen detections with diagnostic multiplex assays. Front Microbiol 8:1210
Cybulski RJ Jr, Bateman AC, Bourassa L, Bryan A, Beail B, Matsumoto J, Cookson BT, Fang FC (2018) Clinical impact of a multiplex gastrointestinal polymerase chain reaction panel in patients with acute gastroenteritis. Clin Infect Dis 67(11):1688–1696
Axelrad JE, Freedberg DE, Whittier S, Greendyke W, Lebwohl B, Green DA (2019) Impact of gastrointestinal panel implementation on health care utilization and outcomes. J Clin Microbiol 57(3):e01775
Hitchcock MM, Gomez CA, Banaei N (2018) Low yield of FilmArray GI panel in hospitalized patients with diarrhea: an opportunity for diagnostic stewardship intervention. J Clin Microbiol 56(3):e01558
Acknowledgments
We acknowledge the excellent technical support of Marjeta Hofko.
Funding
Open Access funding provided by Projekt DEAL.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors A.D. and M.A. have received speaker’s honoraria from Seegene. All the other authors declare that they have no conflict of interest.
Disclaimer
Seegene had no influence on the design of this study, data evaluation, and interpretation and was not involved in manuscript preparation.
Ethical approval/informed consent
This study was done as an observational study analyzing diagnostic procedures with anonymized data for which no ethical approval, and informed consent is required according to the local ethics advisory board of the Heidelberg University Hospital.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zimmermann, S., Horner, S., Altwegg, M. et al. Workflow optimization for syndromic diarrhea diagnosis using the molecular Seegene Allplex™ GI-Bacteria(I) assay. Eur J Clin Microbiol Infect Dis 39, 1245–1250 (2020). https://doi.org/10.1007/s10096-020-03837-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-020-03837-4