Abstract
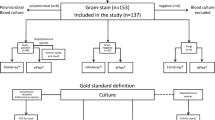
Intra-abdominal infections (IAIs) are one of the most common type of infections in patients with sepsis and an important cause of death in intensive care units. Early detection and treatment are necessary to reduce patient complications and improve outcomes. The Unyvero IAI Application (Curetis GmbH) is the first automated assay to rapidly and simultaneously identify a large panel of bacteria, fungi, toxins, and antibiotic resistance markers directly from IAI-related samples. The assay was evaluated in four European clinical laboratories in comparison to routine microbiological practices. A total of 300 clinical samples were tested with an overall sensitivity of 89.3% and specificity of 99.5%, while time to results was reduced by an average of about 17 h compared to identification (ID) results and 41 h compared to full antibiotic susceptibility testing (AST) results. The Unyvero IAI was able to detect additional microorganisms compared with culture, in particular anaerobes, with most detections confirmed by sequencing. The most frequent resistance markers detected were mecA/mecC (n = 25), aacA4 (n = 20), and blaCTX-M (n = 17) and carbapenemase genes were identified in nine specimens. Further studies are now required to determine the clinical impact of this new rapid test which could play a role in the successful treatment of IAI.

Similar content being viewed by others
References
Claridge JA, Banerjee A, Kelly KB, Leukhardt WH, Carter JW, Haridas M, Malangoni MA (2014) Bacterial species-specific hospital mortality rate for intra-abdominal infections. Surg Infect 15(3):194–199
Labricciosa FM, Sartelli M, Abbo LM, Barbadoro P, Ansaloni L, Coccolini F, Catena F (2018) Epidemiology and risk factors for isolation of multi-drug-resistant organisms in patients with complicated intra-abdominal infections. Surg Infect 19(3):264–272
Hu Y, Yang X, Qin J, Lu N, Cheng G, Wu N et al (2013) Metagenome-wide analysis of antibiotic resistance genes in a large cohort of human gut microbiota. Nat Commun 4:2151
Lawley TD, Walker AW (2013) Intestinal colonization resistance. Immunology 138(1):1–11
Sheng WH, Badal RE, Hsueh PR, SMART program (2013) Distribution of extended-spectrum β-lactamases, AmpC β-lactamases, and carbapenemases among Enterobacteriaceae isolatescausing intra-abdominal infections in the Asia-Pacific region: results of the study for Monitoring Antimicrobial Resistance Trends (SMART). Antimicrob Agents Chemother 57(7):2981–2988
Hayajneh WA, Hajj A, Hulliel F, Sarkis DK, Irani-Hakimeh N, Kazan L, Badal RE (2015) Susceptibility trends and molecular characterization of Gram-negative bacilli associated with urinary tract infection and intra-abdominal infections in Jordan and Lebanon: SMART 2011-2013. Int J Infect Dis 35:56–61
Buss SN, Leber A, Chapin K, Fey PD, Bankowski MJ, Jones MK et al (2015) Multicenter evaluation of the BioFire FilmArray gastrointestinal panel for etiologic diagnosis of infectious gastroenteritis. J Clin Microbiol 53(3):915–925
Khare R, Espy MJ, Cebelinski E, Boxrud D, Sloan LM, Cunningham SA et al (2014) Comparative evaluation of two commercial multiplex panels for detection of gastrointestinal pathogens by use of clinical stool specimens. J Clin Microbiol 52(10):3667–3673
Piralla A, Lunghi G, Ardissino G, Girello A, Premoli M, Bava E et al (2017) FilmArray GI panel performance for the diagnosis of acute gastroenteritis or hemorragic diarrhea. BMC Microbiol 17(1):111
Spina A, Kerr KG, Cormican M, Barbut F, Eigentler A, Zerva L et al (2015) Spectrum of enteropathogens detected by the FilmArray GI panel in a multicentre study of community-acquired gastroenteritis. Clin Microbiol Infect 21(8):719–728
Edelsberg J, Berger A, Schell S, Mallick R, Kuznik A, Oster G (2008) Economic consequences of failure of initial antibiotic therapy in hospitalized adults with complicated intra-abdominal infections. Surg Infect 9(3):335–347
Montravers P, Gauzit R, Muller C, Marmuse JP, Fichelle A, Desmonts JM (1996) Emergence of antibiotic-resistant bacteria in cases of peritonitis after intraabdominal surgery affects the efficacy of empirical antimicrobial therapy. Clin Infect Dis 23(3):486–494
Mosdell DM, Morris DM, Voltura A, Pitcher DE, Twiest MW, Milne RL et al (1991) Antibiotic treatment for surgical peritonitis. Ann Surg 214(5):543–549
Montravers P, Dupont H, Leone M, Constantin JM, Mertes PM, Société française d’anesthésie et de réanimation (Sfar) et al (2015b) Guidelines for management of intra-abdominal infections. Anaesth Crit Care Pain Med 34(2):117–130
Solomkin JS, Mazuski JE, Bradley JS, Rodvold KA, Goldstein EJ, Baron EJ et al (2010) Diagnosis and management of complicated intra-abdominal infection in adults and children: guidelines by the Surgical Infection Society and the Infectious Diseases Society of America. Clin Infect Dis 50(2):133–164
Montravers P, Dufour G, Guglielminotti J, Desmard M, Muller C, Houissa H et al (2015a) Dynamic changes of microbial flora and therapeutic consequences in persistent peritonitis. Crit Care 19:70
Orsi GB, Ciorba V (2013) Vancomycin resistant enterococci healthcare associated infections. Ann Ig 25(6):485–492
Cattoir V, Giard JC (2014) Antibiotic resistance in Enterococcus faecium clinical isolates. Expert Rev Anti-Infect Ther 12(2):239–248
Montravers P, Dupont H, Gauzit R, Veber B, Auboyer C, Blin P et al (2006) Candida as a risk factor for mortality in peritonitis. Crit Care Med 34(3):646–652
Zautner AE, Gross U, Emele MF, Hagen RM, Frickmann H (2017) More pathogenicity or just more pathogens?-on the interpretation problem of multiple pathogen detections with diagnostic multiplex assays. Front Microbiol 8:1210
Acknowledgements
Curetis GmbH provided the panel reagents and instruments used in this study, and performed the additional PCR and sequencing for discrepant results analysis. All authors have reviewed and agreed to this version of the manuscript.
H. Ciesielczuk presented some of this data at ECCMID 2017.
Funding
All study reagents, consumables, and costs were provided by Curetis GmbH.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Informed consent
Informed consent was not required for this study as there were no human participants.
Appendix
Appendix
Microbiology culture testing. The specimens were tested for pathogens using the standard procedure of each laboratory as described below.
Barts Health NHS Trust, London: Gram stain was performed on all sample types except drain fluids. Cell counts were performed on all ascitic fluids and peritoneal fluids and both specimens were concentrated prior to subculture on solid agar and incubation in BacT Alert (BioMerieux) blood culture bottles for 4 days. All sample types were cultured microaerophilically on blood agar at 37 °C for 48 h and also anaerobically on blood agar with a metronidazole disc (5 μg) for 48 h. Bile samples were also cultured on XLD agar (Xylose-Lysine-Desoxycholate) and incubated at 37 °C in O2 for 24 h. If Actinomyces or Nocardia species were suspected, pus samples were cultured on FBH agar and incubated anaerobically for 10 days. Tissues were transferred (1 cm3) into 5 ml saline containing glass ballotini beads and vortexed at high speed for 10 s. The bead mill was then subcultured as described above, with the addition of a chocolate agar plate if indicated by the Gram stain result. All isolates were identified by MALDI-TOF MS (Bruker) and antibiotic susceptibility testing was performed on the MicroScan (Beckman Coulter), with the exception of streptococci, enterococci and anaerobes which underwent disc diffusion according to BSAC guidelines.
Toulouse University Hospital Center: Gram stain was performed on all sample types. Peritoneal fluids and bile were inoculated onto blood agar, chocolate agar (incubated microaerophilically), Colombia CAN agar, BromoCresol Purple agar, Sabouraud agar (incubated aerobically), and Wilkins-Chalgren agar (incubated anaerobically). Peritoneal fluids were also inoculated into blood culture bottles and subcultured to blood agar and BromCresol Purple agar when they became positive. For ascitic fluids, pancreatic fluids, and abscesses, samples were inoculated onto blood agar, and chocolate agar, (incubated microaerophilically), Sabouraud agar (incubated aerobically), blood agar (incubated anaerobically), and thioglycolate medium. Agar plates were read daily for up to 5 days and colonies was identified using MALDI-TOF MS (Bruker). Susceptibility testing was performed using the Vitek 2 system for Gram negative rods, Enterococcus species and staphylococci, with streptococci and anaerobes tested by disk diffusion according to the recommendations of the CA-SFM/EUCAST (Comité de l’Antibiogramme de la Société Française de Microbiologie/European Committee on Antimicrobial Susceptibility Testing).
Amiens University Hospital Center: Gram staining was performed on pus samples only. IAI samples were cultured onto blood agar under aerobic and anaerobic conditions; Schaedler Neo Vanco + 5% sheep blood (SNVS) agar and colistin-nalidixic acid (CNA) agar under anaerobic conditions; chocolate culture medium under microaerophilic conditions; and BHI broth under aerobic conditions. Plates were read at 24 h, 48 h, 72 h, and 96 h and colonies identified using MALDI-TOF MS (Bruker). Susceptibility testing was performed by agar diffusion following the CASFM 2013 recommendations.
The Great Romagna Hub Laboratory, Pievesestina: sterile fluids (ascitic, peritoneal, and biliary fluids and abdominal drainages) were collected in 4-mL heparinized tubes and routinely subcultured onto Columbia CNA agar + 5% sheep blood, chocolate agar PolyViteX, mannitol salt 2 agar, Mac Conkey agar, and Candida agar plus brain-heart infusion broth (BHI) at 37 °C. Gram staining was performed on all sample types. If anaerobic bacteria were suspected, the sample was cultured on three additional plates: Columbia agar + 5% sheep blood, Schaedler agar + 5% sheep blood, SNVS (BioMerieux). Plates were read at 24 h and 48 h (if cultured under aerobic conditions) or at 48 h and 72 h if incubated anaerobically. Turbid BHI tubes were subcultured only when solid plates showed no growth. Identification and susceptibility testing were performed using a Vitek2 instrument (BioMérieux). Additionally, a set of blood culture bottles were inoculated for each specimen and incubated at 37 °C for up to 5 days, with appropriate sub-culture depending on whether the aerobic or anaerobic bottle became positive.
Rights and permissions
About this article
Cite this article
Ciesielczuk, H., Wilks, M., Castelain, S. et al. Multicenter performance evaluation of the Unyvero IAI cartridge for detection of intra-abdominal infections. Eur J Clin Microbiol Infect Dis 37, 2107–2115 (2018). https://doi.org/10.1007/s10096-018-3345-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-018-3345-0