Abstract
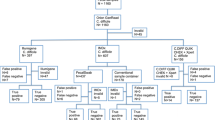
In this study, the usability and performance of GenomEra™ C. difficile and BD Max™ Cdiff nucleic acid amplification tests (NAATs) for the detection of toxigenic Clostridium difficile were investigated in comparison with toxigenic culture and C. difficile toxin A- and toxin B-detecting immunochromatographic antigen (IA) test, the Tox A/B QuikChek®. In total, 302 faecal specimens were collected, 113 of which were in parallel to conventional sample containers and FecalSwab liquid-based microbiology (LBM) tubes. Seventy-nine specimens were considered true-positives for toxigenic C. difficile. The sensitivity and specificity were 97.5 % and 99.6 % and 93.7 % and 98.7 % for the GenomEra and BD Max assays respectively. Toxigenic culture and Tox A/B QuikChek had sensitivity and specificity of 91.1 % and 100 % and 34.2 % and 100 % respectively. Hands-on time for analysing 1 to 24 specimens using NAATs was 1 to 15 min. The rate of PCR inhibition was 0 % for both NAATs with faeces in LBM tubes, while with faeces in conventional sample containers the respective inhibition rates were 5.3 % and 4.4 % for the GenomEra and the BD Max assays. The NAATs demonstrated an excellent analytical performance, reducing significantly the overall workload of laboratory personnel compared with culture and IA test.
Similar content being viewed by others
References
Bartlett JG, Gerding DN (2008) Clinical recognition and diagnosis of Clostridium difficile infection. Clin Infect Dis 46:S12–S18. doi:10.1086/521863
Karas JA, Enoch DA, Aliyu SH (2010) A review of mortality due to Clostridium difficile infection. J Infect 61:1–8. doi:10.1016/j.jinf.2010.03.025
Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP (2010) The role of toxin A and toxin B in Clostridium difficile infection. Nature 467:711–713. doi:10.1038/nature09397
Lyras D, O’Connor JR, Howarth PM, Sambol SP, Carter GP, Phumoonna T, Poon R, Adams V, Vedantam G, Johnson S, Gerding DN, Rood JI (2009) Toxin B is essential for virulence of Clostridium difficile. Nature 458:1176–1179. doi:10.1038/nature07822
Elliott B, Squire MM, Thean S, Chang BJ, Brazier JS, Rupnik M, Riley TV (2011) New types of toxin A-negative, toxin B-positive strains among clinical isolates of Clostridium difficile in Australia. J Med Microbiol 60:1108–1111. doi:10.1099/jmm. 0.031062-0
Warny M, Pepin J, Fang A, Killgore G, Thompson A, Brazier J, Frost E, McDonald LC (2005) Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079–1084. doi:10.1016/S0140-6736(05)67420-X
Stewart DB, Berg A, Hegarty J (2013) Predicting recurrence of C. difficile colitis using bacterial virulence factors: binary toxin is the key. J Gastrointest Surg 17:118–124. doi:10.1007/s11605-012-2056-6
Dodek PM, Norena M, Ayas NT, Romney M, Wong H (2013) Length of stay and mortality due to Clostridium difficile infection acquired in the intensive care unit. J Crit Care 28:335–340. doi:10.1016/j.jcrc.2012.11.008
Arroyo LG, Rousseau J, Willey BM, Low DE, Staempfli H, McGeer A, Weese JS (2005) Use of a selective enrichment broth to recover Clostridium difficile from stool swabs stored under different conditions. J Clin Microbiol 43:5341–5343. doi:10.1128/JCM. 43.10.5341-5343.2005
Bliss DZ, Johnson S, Clabots CR, Savik K, Gerding DN (1997) Comparison of cycloserine-cefoxitin-fructose agar (CCFA) and taurocholate-CCFA for recovery of Clostridium difficile during surveillance of hospitalized patients. Diagn Microbiol Infect Dis 29:1–4
Clabots CR, Gerding SJ, Olson MM, Peterson LR, Gerding DN (1989) Detection of asymptomatic Clostridium difficile carriage by an alcohol shock procedure. J Clin Microbiol 27:2386–2387
Marler LM, Siders JA, Wolters LC, Pettigrew Y, Skitt BL, Allen SD (1992) Comparison of five cultural procedures for isolation of Clostridium difficile from stools. J Clin Microbiol 30:514–516
Brazier JS (1998) The diagnosis of Clostridium difficile-associated disease. J Antimicrob Chemother 41:29–40
Bruins MJ, Verbeek E, Wallinga JA, Bruijnesteijn van Coppenraet LE, Kuijper EJ, Bloembergen P (2012) Evaluation of three enzyme immunoassays and a loop-mediated isothermal amplification test for the laboratory diagnosis of Clostridium difficile infection. Eur J Clin Microbiol Infect Dis 31:3035–3039. doi:10.1007/s10096-012-1658-y
Culbreath K, Ager E, Nemeyer RJ, Kerr A, Gilligan PH (2012) Evolution of testing algorithms at a university hospital for detection of Clostridium difficile infections. J Clin Microbiol 50:3073–3076. doi:10.1128/JCM. 00992-12
Walkty A, Lagacé-Wiens PR, Manickam K, Adam H, Pieroni P, Hoban D, Karlowsky JA, Alfa M (2013) Laboratory diagnosis of Clostridium difficile infection—evaluation of an algorithmic approach in comparison with the Illumigene® assay. J Clin Microbiol 51:1152–1157. doi:10.1128/JCM. 03203-12
Alonso R, Muñoz C, Peláez T, Cercenado E, Rodríguez-Creixems M, Bouza E (1997) Rapid detection of toxigenic Clostridium difficile strains by a nested PCR of the toxin B gene. Clin Microbiol Infect 3:145–147
Bélanger SD, Boissinot M, Clairoux N, Picard FJ, Bergeron MG (2003) Rapid detection of Clostridium difficile in feces by real-time PCR. J Clin Microbiol 41:730–734. doi:10.1128/JCM. 41.2.730-734.2003
Kato H, Kato N, Watanabe K, Iwai N, Nakamura H, Yamamoto T, Suzuki K, Kim SM, Chong Y, Wasito EB (1998) Identification of toxin A-negative, toxin B-positive Clostridium difficile by PCR. J Clin Microbiol 36:2178–2182
Dubberke ER, Han Z, Bobo L, Hink T, Lawrence B, Copper S, Hoppe-Bauer J, Burnham CA, Dunne WM Jr (2011) Impact of clinical symptoms on interpretation of diagnostic assays for Clostridium difficile infections. J Clin Microbiol 49:2887–2893. doi:10.1128/JCM. 00891-11
Eastwood K, Else P, Charlett A, Wilcox M (2009) Comparison of nine commercially available Clostridium difficile toxin detection assays, a real-time PCR assay for C. difficile tcdB, and a glutamate dehydrogenase detection assay to cytotoxin testing and cytotoxigenic culture methods. J Clin Microbiol 47:3211–3217. doi:10.1128/JCM. 01082-09
Buchan BW, Mackey TL, Daly JA, Alger G, Denys GA, Peterson LR, Kehl SC, Ledeboer NA (2012) Multicenter clinical evaluation of the portrait toxigenic C. difficile assay for detection of toxigenic Clostridium difficile strains in clinical stool specimens. J Clin Microbiol 50:3932–3936. doi:10.1128/JCM. 02083-12
Chapin KC, Dickenson RA, Wu F, Andrea SB (2011) Comparison of five assays for detection of Clostridium difficile toxin. J Mol Diagn 13:395–400. doi:10.1016/j.jmoldx.2011.03.004
Le Guern R, Herwegh S, Grandbastien B, Courcol R, Wallet F (2012) Evaluation of a new molecular test, the BD Max Cdiff, for detection of toxigenic Clostridium difficile in fecal samples. J Clin Microbiol 50:3089–3090. doi:10.1128/JCM. 01250-12
Shin BM, Mun SJ, Yoo SJ, Kuak EY (2012) Comparison of BD GeneOhm Cdiff and Seegene Seeplex ACE PCR assays using toxigenic Clostridium difficile culture for direct detection of tcdB from stool specimens. J Clin Microbiol 50:3765–3767. doi:10.1128/JCM. 01440-12
Terhes G, Urbán E, Sóki J, Nacsa E, Nagy E (2009) Comparison of a rapid molecular method, the BD GeneOhm Cdiff assay, to the most frequently used laboratory tests for detection of toxin-producing Clostridium difficile in diarrheal feces. J Clin Microbiol 47:3478–3481. doi:10.1128/JCM. 01133-09
Hirvonen JJ, Mentula S, Kaukoranta S-S (2013) Evaluation of a new automated homogeneous PCR assay, GenomEra C. difficile, for rapid detection of toxigenic Clostridium difficile in fecal specimens. J Clin Microbiol 51:2908–2912. doi:10.1128/JCM. 01083-13
Stellrecht KA, Espino AA, Maceira VP, Nattanmai SM, Butt SA, Wroblewski D, Hannett GE, Musser KA (2014) Premarket evaluations of the IMDx C. difficile for Abbott m2000 assay and the BD Max Cdiff assay. J Clin Microbiol 52:1423–1428. doi:10.1128/JCM. 03293-13
Quinn CD, Sefers SE, Babiker W, He Y, Alcabasa R, Stratton CW, Carroll KC, Tang Y-W (2010) C. Diff QuikChek Complete enzyme immunoassay provides a reliable first-line method for detection of Clostridium difficile in stool specimens. J Clin Microbiol 48:603–605. doi:10.1128/JCM. 01614-09
Crobach MJ, Dekkers OM, Wilcox MH, Kuijper EJ (2009) European Society of Clinical Microbiology and Infectious Diseases (ESCMID): data review and recommendations for diagnosing Clostridium difficile-infection (CDI). Clin Microbiol Infect 15:1053–1066. doi:10.1111/j.1469-0691.2009.03098.x
Brecher SM, Novak-Weekley SM, Nagy E (2013) Laboratory diagnosis of Clostridium difficile infections: there is light at the end of the colon. Clin Infect Dis 57:1175–1181. doi:10.1093/cid/cit424
Rousseau C, Poilane I, De Pontual L, Maherault AC, Le Monnier A, Collignon A (2012) Clostridium difficile carriage in healthy infants in the community: a potential reservoir for pathogenic strains. Clin Infect Dis 55:1209–1215. doi:10.1093/cid/cis637
Matsuki S, Ozaki E, Shozu M, Inoue M, Shimizu S, Yamaguchi N, Karasawa T, Yamagishi T, Nakamura S (2005) Colonization by Clostridium difficile of neonates in a hospital, and infants and children in three day-care facilities of Kanazawa, Japan. Int Microbiol 8:43–48
Kyne L, Warny M, Qamar A, Kelly CP (2000) Asymptomatic carriage of Clostridium difficile and serum levels of IgG antibody against toxin A. N Engl J Med 342:390–397
Riggs MM, Sethi AK, Zabarsky TF, Eckstein EC, Jump RL, Donskey CJ (2007) Asymptomatic carriers are a potential source for transmission of epidemic and nonepidemic Clostridium difficile strains among long-term care facility residents. Clin Infect Dis 45:992–998. doi:10.1086/521854
Pasternack R, Vuorinen P, Kuukankorpi A, Pitkäjärvi T, Miettinen A (1996) Detection of Chlamydia trachomatis infections in women by Amplicor PCR: comparison of diagnostic performance with urine and cervical specimens. J Clin Microbiol 34:995–998
Hirvonen JJ, Kaukoranta SS (2014) Comparison of FecalSwab and ESwab devices for storage and transportation of diarrheagenic bacteria. J Clin Microbiol 52:2334–2339. doi:10.1128/JCM. 00539-14
Acknowledgements
Nancy Lahtinen, Mikaela Eur, and Marianne Nynäs are gratefully acknowledged for their help in sample preparations and workload analysis. We have no conflicts of interests to declare.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hirvonen, J.J., Kaukoranta, SS. Comparison of BD Max Cdiff and GenomEra C. difficile molecular assays for detection of toxigenic Clostridium difficile from stools in conventional sample containers and in FecalSwabs. Eur J Clin Microbiol Infect Dis 34, 1005–1009 (2015). https://doi.org/10.1007/s10096-015-2320-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-015-2320-2