Abstract
The imipenem and meropenem-resistant strains Citrobacter freundii HS70 and Escherichia coli HS510 were isolated from patients in Shanghai, China. By isoelectric focusing, PCR amplification and sequencing, these strains were each found to produce four β-lactamases: TEM-1, KPC-3, SHV-7 and CTX-M-14. A conjugation experiment and plasmid restriction digestion revealed that the bla KPC-3 gene was located on the same plasmid in both isolates. Bidirectional primer walking sequencing showed that the nucleotide sequence surrounding the 3.8 kb bla KPC-3 contained a 671-bp insertion similar to that previously characterized in China. The insertion was located between the promoter and the coding region of the bla KPC-3 gene. Susceptibility testing performed on recombinant strains carrying the bla KPC-3 gene with or without the insertion revealed that minimum inhibitory concentrations of imipenem, meropenem, cefepime, and cefotaxime for E. coli EMU-KPC3 (without insertion) were four times higher than that of E. coli EKPC3 (with insertion). The 671 bp insertion reduced bla KPC-3 expression significantly. Taken together, these results suggest that KPC-3-producing C. freundii and E. coli have begun to emerge in our hospital.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The extensive use of carbapenems has resulted in the emergence of carbapenem-resistant Enterobacteriaceae strains [4]. The resistance may be mediated by the production of carbapenemases [15, 19], as well as AmpC hyper-production combined with decreased outer membrane permeability due to loss or alteration of porins [14]. The Ambler class A Klebsiella pneumoniae carbapenemase (KPC) enzymes [13] are able to hydrolyze all known β-lactam-containing molecules and are the most frequently observed class A carbapenemases.
KPC-1, a plasmid encoded β-lactamase, was first identified from K. pneumoniae in North Carolina (USA) and is identical to the bla KPC-2 gene by sequencing [25, 26]. Strains harboring bla KPC-1/KPC-2 have also been isolated from patients in France [9], Israel [12], South America [18], Greece [3], and China [20]. The bla KPC-3 sequence (GenBank AM774409) is found in the same genetic environment as bla KPC-2 in Salmonella cubana 4707 (GenBank AF481906) [5], and this KPC enzyme is now prevalent in America [22], Israel [6], and the United Kingdom [23].
In this study, we identified strains of Citrobacter freundii and E. coli isolated from Chinese patients that express KPC-3. The background of the bla KPC-3 gene was different from that reported outside of China and it was similar to that reported in China previously [27].
Materials and methods
Bacterial strains and plasmids
The imipenem and meropenem-resistant strain C. freundii HS70 was isolated from the urine of a 53-year-old female patient hospitalized in Huashan Hospital, Fudan University. The imipenem and meropenem-resistant strain E. coli HS510 was isolated from urine of a 59-year-old female inpatient at the same hospital. Both strains were identified by Vitek-32 (BioMerieux, Marcy, France). E. coli J53 was used as a recipient in conjugal mating experiments, whereas E. coli DH5α was used for cloning. Other derivative strains and plasmids used in this study are listed in Table 1.
Antimicrobial susceptibility testing
Minimum inhibitory concentrations (MICs) for organisms were determined by the Mueller-Hinton (M-H) agar dilution method according to guidelines of the Clinical and Laboratory Standards Institute [2]. Antimicrobial agents evaluated included imipenem, meropenem, cefepime, cefotaxime, ampicillin, ciprofloxacin, and gentamicin. All were obtained from Oxoid (Basingstoke, England). E. coli ATCC25922 was used for quality control.
Conjugation experiments and plasmid restriction enzyme digestion analysis
Transfer of imipenem resistance was studied by performing conjugation experiments as previously described [24] with E. coli J53 as the recipient. Transconjugants were selected from agar plates supplemented with sodium azide (100 μg/mL; Oxoid, Basingstoke, England) and ceftazidime (2 μg/mL; Oxoid, Basingstoke, England), and identified by VITEK-32. For the plasmid restriction enzyme analysis, XbaI and ClaI (Takara, Dalian, China) were used. Digested plasmid DNA samples from transconjugants were then analyzed by electrophoresis on 0.6% agarose gels at a constant voltage of 100 V for 0.5 h.
Isoelectric focusing of β-lactamases
Crude cell lysates were prepared by a previously described freeze–thaw procedure [17]. Isoelectric focusing was performed as described by Matthew and Harris [8]. Cell extracts were loaded onto prepared polyacrylamide gel plates (pH 3 to 9; Amersham Biosciences, Uppsala, Sweden) and electrophoresed to equilibrium using Pharmacia PhastSystem (Uppsala, Sweden). β-lactamases were then visualized by staining the gel with a 0.05% solution of nitrocefin (BD Biosciences, San Jose, CA, USA). The isoelectric points of TEM-1, KPC-3, SHV-7 and CTX-M-14 were determined by comparison to known pIs of the β-lactamases (TEM-12, pI 5.25; TEM-28, pI 6.1; SHV-7, pI 7.6; and ACT-1, pI 9.0).
PCR analysis and nucleotide sequencing
Crude genomic DNA was extracted from the isolates by heat lysis. β-lactamase genes were identified by PCR with specific primers designed to sequences of known β-lactamase genes, including bla TEM, bla SHV, bla KPC, bla CTX-M-1, bla CTX-M-9, and bla CTX-M-2. For PCRs the LA Taq DNA polymerase (Takara, Dalian, China) was used according to the manufacturer’s instructions. Primer sequences are listed in Table 2. PCR amplifications were performed and PCR products were then sequenced by an ABI 3730 analyzer, and the obtained sequences were aligned with sequence data from GenBank.
Analysis of the genetic environment of the blaKPC-3 gene and plasmid construction
The genetic context of the bla KPC-3 gene was examined by bidirectional primer walking sequencing, performed on plasmids from transconjugants using primers designed previously [20]. The obtained sequence was aligned with β-lactamase sequences from the GenBank database using the BLAST program. Primers KPC-3 F and KPC-3R were used to amplify the DNA fragments, which included the entire bla KPC gene and the region flanking the insertion in the plasmid isolated from the Chinese patients. The obtained PCR products and plasmid pACYC184 were digested using Bam HI and EcoRI, then ligated using T4 DNA ligase. The obtained plasmid pACYC184-KPC3 was electroporated into E. coli DH5α. Clones were selected on Luria-Bertani agar plates containing chloramphenicol (40 mg/mL) and imipenem (1.5 mg/mL). Primers MU-KPC-3F and MU-KPC-3R were designed to amplify the whole recombinant plasmid pACYC184-KPC3 without the 671 bp insertion fragment. The obtained PCR products were ligated by using a MutantBEST kit (Takara) to produce the plasmid pMU-ACYC184-KPC3. The recombinant plasmids pACYC184-KPC3 (with insertion) and pMU-ACYC184-KPC3 (without insertion) were then individually transformed into E. coli DH5α by the calcium phosphate method.
Results
Antimicrobial resistance
E. coli HS510 was isolated from the urine of a 59-year-old female patient and was found resistant to imipenem and meropenem, with a MIC of 16 μg/mL for both antibiotics. C. freundii HS70 was isolated from the urine of a 53-year-old female patient and had a MIC of ≥128 μg/mL for both antibiotics (Table 3).
Isoelectric focusing and PCR analysis of β-lactamases
In order to understand the antimicrobial resistant phenotype, β-lactamases were analyzed by isoelectric focusing, PCR and PCR product sequencing. Isoelectric focusing of clinical isolates revealed that both E. coli HS510 and C. freundii HS70 produced four β-lactamases which possess pIs of 5.4, 6.7, 7.6, and 8.1 (Fig. 1), respectively. To determine which lactamases they were, PCR was performed on DNA from the clinical isolates with primers specific for KPC, TEM, SHV, CTX-M-1, CTX-M-2, and CTX-M-9. Sequencing analysis of two clinical isolates confirmed that both E. coli HS510 and C. freundii HS70 carried bla TEM-1, bla CTX-M-14, bla SHV-7 and bla KPC-3.
Isoelectric focusing patterns of clinical isolates E. coli HS510 and C. freundii HS70. Lane 1: Cell lysate prepared from C. freundii HS70 producing TEM-1 (pI 5.4), KPC-3 (pI 6.7), SHV-12 (pI 7.6), and CTX-M-14 (pI 7.9). Lane 2: Cell lysate prepared from E. coli HS510 producing TEM-1, KPC-3, SHV-12 and CTX-M-14
Plasmid profile analysis
Conjugation experiments successfully transferred a plasmid from both clinical isolates to the recipient E. coli J53. In order to understand which resistant genes can be transferred by plasmid conjugation, we used PCR to identify every β-lactamase gene that the two clinical isolates contained on transconjugants. PCR results confirmed that both transconjugants only carried the blaKPC-3 gene. Restriction enzyme (XbaI and ClaI) digestion of plasmids from transconjugants showed that they had identical enzyme digestion maps (Fig. 2). Our results indicated that the bla KPC gene was located on the same plasmid in both clinical isolates.
Characterization of the genetic environment of the blaKPC-3 gene
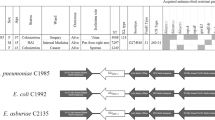
Bidirectional primer walking performed on transconjugated plasmids produced a 3,850 bp fragment. The nucleotide sequence was assigned GenBank accession number FJ609231. Sequence alignment showed that the FJ609231 sequence was similar to AM774409.1. FJ609231 carried the bla KPC-3 gene; EU176014.1 and FJ628167 each carried a bla KPC-2 gene. Furthermore, FJ609231 showed a similar structure and context with FJ628167, except for a 671-bp fragment insertion between the bla KPC-3 promoter and the bla KPC coding region (between positions 20571 and 20572 of FJ628167; see Fig. 3). As for FJ628167, an ISKpn6-like element is located downstream of the bla KPC-3 gene and a Tn3 transposon and ISKpn8 is located upstream of the bla KPC-3 gene [27]. The additional 671 bp insertion looks like a piece of TEM1. The nucleotide sequence area is from 982 to 1928 of FJ609231, and looks like a rearranged FJ223605.1.
Schematic representation of novel genetic structure involved in bla KPC-3 gene found in bacteria isolated from Chinese patients. EU176014.1 contains ISKpn7, bla KPC-3 and ISKpn6; AM774409.1 contains ISKpn7, bla KPC-2 and ISKpn6; FJ628167 contains ISKpn8, bla KPC-2 and an ISKpn6-like element; this study contained ISKpn8, truncated β-latamase, bla KPC-3 and an ISKpn6-like element
Antimicrobial susceptibility testing of recombinants
To investigate the effects of the inserted 671-bp fragment on bla KPC-3 expression, plasmids pKPC3-184 and pMU-KPC3-184 were constructed using the vector pACYC184. The plasmid pKPC3-184 was derived from plasmid pHS70, and pMU-KPC3-184 was derived from plasmid pKPC3-184. The genetic environment of the bla KPC gene on pKPC3-184 was the same with pHS70. The genetic environment of the bla KPC gene on pMU-KPC3-184 was the same with pHS70 except for a 671-bp fragment insertion between the bla KPC-3 promoter and the bla KPC coding region. Recombinants E. coli EKPC3 and E. coli EMU-KPC3 (with plasmids pKPC3-184 and pMU-KPC3-184, respectively) were constructed and tested for antimicrobial susceptibility. The MICs of E. coli EMU-KPC3 for imipenem and meropenem were four times higher than that of E. coli EKPC3 (32 μg/mL vs. 8 μg/mL) as were MICs for cefepime and cefotaxime (64 μg/mL for E. coli EMU-KPC3 vs. 16 μg/mL for E. coli EKPC3), implying that the 671-bp fragment insertion decreased KPC-3 expression.
Discussion
This is the first time that the bla KPC-3 gene was reported in China. Nine KPC variants have so far been described (KPC-2 to KPC-10; KPC-1 and KPC-2 are identical) in different parts of the world; variants of KPC-1/2 differ by, at most, two amino acid substitutions [1, 5, 6, 16, 21, 22]. The emergence of KPC-type carbapenem-hydrolyzing enzymes is alarming. E. coli transconjugants carrying the bla KPC-3 gene from either clinical isolate in our study had MICs of 2 μg/mL for both imipenem and meropenem. Additional mechanisms must underlie the drug resistance of clinical isolates C. freundii HS70 and E. coli HS510 since their MICs for imipenem and meropenem were greater than that of their E. coli transconjugants. Previous reports showed that the lost porins OmpK36 and OmpK35 are perhaps associated with the increase in MICs for carbapenems [22].
Our conjugation experiments, plasmid restriction enzyme digestion analysis and PCR of the β-lactamase gene confirmed that bla KPC-3 from both clinical isolates is located on the same conjugational plasmid. Sequence analysis revealed that 3.8 Kb of DNA sequence surrounding the bla KPC-3 gene of these two isolates was also the same, and its construction was similar to AM774409.1 from Enterobacter cloacae and EU176014.1 from Klebsiella pneumoniae. It was composed of ISKpn8, the bla KPC-3 promoter, a 671-bp insertion, the bla KPC-3 gene and an ISkpn6-like element (Fig. 3). This DNA fragment is one part of a Tn3-based transposon, Tn4401, which is likely to be the origin of bla KPC mobilization and further insertion into various plasmids of non-clonally related organisms [10].
This report provides further confirmation of the putatively transposable element found on plasmids of K. pneumoniae and P. aeruginosa that may be responsible for the rapid global dissemination of this gene. The region upstream of the bla KPC-3 identified here differs from AM774409.1 and EU176014.1 (Fig. 3). A 671-bp fragment insertion was found between the bla KPC-3 promoter and the bla KPC-3 coding region (between positions 20571 and 20572 of FJ628167). This finding confirms Naas’s proposition [10] that this region of the element is unstable, and suggests that other isoforms of Tn4401 do exist. Naas et al. identified a 100-bp deletion upstream of bla KPC from two clinical strains of K. pneumonia, namely, GR and YC [10].
Interestingly, we found that the insertion decreased bla KPC-3 expression. Our results provide important information on the genetic environment of bla KPC-3 genes identified in clinical isolates in China and the significance of bla KPC-3 in this genetic context warrants further study.
References
Alba J, Ishii Y, Thomson K, Moland ES, Yamaguchi K (2005) Kinetics study of KPC-3, a plasmid-encoded class A carbapenem-hydrolyzing β-lactamase. Antimicrob Agents Chemother 49:4760–4762
Clinical and Laboratory Standards Institute (2009) Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 7th ed. Approved standard M7–A8, Wayne, PA, USA
Cuzon G, Naas T, Demachy MC, Nordmann P (2008) Plasmid-mediated carbapenem-hydrolyzing β-lactamase KPC-2 in Klebsiella pneumoniae isolate from Greece. Antimicrob Agents Chemother 52:796–797
Deshpande LM, Jones RN, Fritsche TR, Sader HS (2006) Occurrence and characterization of carbapenemase-producing Enterobacteriaceae: report from the SENTRY Antimicrobial Surveillance Program (2000–2004). Microb Drug Resist 12:223–230
Dortet L, Radu I, Gautier V, Blot F, Chachaty E, Arlet G (2008) Intercontinental travels of patients and dissemination of plasmid-mediated carbapenemase KPC-3 associated with OXA-9 and TEM-1. J Antimicrob Chemother 61:455–457
Leavitt A, Navon-Venezia S, Chmelnitsky I, Schwaber MJ, Carmeli Y (2007) Emergence of KPC-2 and KPC-3 in carbapenem-resistant Klebsiella pneumoniae strains in an Israeli hospital. Antimicrob Agents Chemother 51:3026–3029
Kitchel B, Rasheed JK, Patel JB, Srinivasan A, Navon-Venezia S, Carmeli Y, Brolund A, Giske CG (2009) Molecular epidemiology of KPC-producing Klebsiella pneumoniae isolates in the United States: clonal expansion of multilocus sequence type 258. Antimicrob Agents Chemother 53:3365–3370
Matthew M, Harris AM (1976) Identification of β-lactamases by analytical isoelectric focusing: correlation with bacterial taxonomy. J Gen Microbiol 94:55–67
Naas TP, Nordmann P, Vedel G, Poyart C (2005) Plasmid-mediated carbapenem-hydrolyzing β-Lactamase KPC in a Klebsiella pneumoniae isolate from France. Antimicrob Agents Chemother 49:4423–4424
Naas T, Cuzon G, Villegas MV, Lartigue MF, Quinn JP, Nordmann P (2008) Genetic structures at the origin of acquisition of the β-lactamase bla KPC gene. Antimicrob Agents Chemother 52:1257–1263
Navon-Venezia S, Leavitt AA, Schwaber MJ, Rasheed JK, Srinivasan A, Patel JB, Carmeli Y, Israeli KPC, Kpn Study Group (2009) First report on a hyperepidemic clone of KPC-3-producing Klebsiella pneumoniae in Israel genetically related to a strain causing outbreaks in the United States. Antimicrob Agents Chemother 53:818–820
Navon-Venezia S, Chmelnitsky I, Leavitt A, Schwaber MJ, Schwartz D, Carmeli Y (2006) Plasmid-mediated imipenem-hydrolyzing enzyme KPC-2 among multiple carbapenem-resistant Escherichia coli clones in Israel. Antimicrob Agents Chemother 50:3098–3101
Nordann P, Cuzon G, Naas T (2009) The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis 9:228–236
Poirel L, Heritier C, Spicq C, Nordmann P (2004) In vivo acquisition of high-level resistance to imipenem in Escherichia coli. J Clin Microbiol 42:3831–3833
Queenan AM, Bush K (2007) Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev 20:440–458
Robledo IE, Moland ES, Aquino EA, Vázquez GJ, Santé MI, Bertrán J, Hanson ND (2007) First report of a KPC-4 and CTX-M producing K. pneumoniae (Kp) isolated from Puerto Rico (PR). Abstracts of the 47th Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, IL, Washington, DC, USA. American Society for Microbiology, p 142, Abstract C2-1933
Sykes RB, Bonner DP, Bush K, Georgopapadakou NH (1982) Azthreonam (SQ 26, 776), a synthetic monobactam specifically active against aerobic gram-negative bacteria. Antimicrob Agents Chemother 21:85–92
Villegas MV, Lolans K, Correa A, Suarez CJ, Lopez JA, Vallejo M, Quinn JP, Colombian Nosocomial Resistance Study Group (2006) First detection of the plasmid-mediated class A carbapenemase KPC-2 in clinical isolates of Klebsiella pneumoniae from South America. Antimicrob Agents Chemother 50:2880–2882
Walsh TR (2005) The emergence and implications of metallo-lactamases in Gram-negative bacteria. Clin Microbiol Infect 11(suppl 6):2–9
Wei ZQ, Du XX, Yu YS, Shen P, Chen YG, Li LJ (2007) Plasmid-mediated KPC-2 in a Klebsiella pneumoniae isolate from China. Antimicrob Agents Chemother 51:763–765
Wolter DJ, Kurpiel PM, Woodford N, Palepou MF, Goering RV, Hanson ND (2009) Phenotypic and enzymatic comparative analysis between the novel KPC variant, KPC-5, and its evolutionary variants, KPC-2 and KPC-4. Antimicrob Agents Chemother 53:557–562
Woodford N, Tierno PM Jr, Young K, Tysall L, Palepou MF, Ward E, Painter RE, Suber DF, Shungu D, Silver LL, Inglima K, Kornblum J, Livermore DM (2004) Outbreak of Klebsiella pneumoniae producing a new carbapenem-hydrolyzing class A β-lactamase, KPC-3, in a New York Medical Center. Antimicrob Agents Chemother 48:4793–4799
Woodford N, Zhang J, Warner M, Kaufmann ME, Matos J, MacDonald A, Brudney D, Sompolinsky D, Navon-Venezia S, Livermore DM (2008) Arrival of Klebsiella pneumoniae producing KPC carbapenemase in the United Kingdom. J Antimicrob Chemother 62:1261–1264
Yan JJ, Wu SM, Tsai SH, Wu JJ, Su IJ (2000) Prevalence of SHV-12 among clinical isolates of Klebsiella pneumoniae producing extended-spectrum β-lactamases and identification of a novel AmpC enzyme (CMY-8) in Southern Taiwan. Antimicrob Agents Chemother 44:1438–1442
Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC (2001) Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 45:1151–1161
Yigit H, Queenan AM, Anderson GJ, Domenech-Sanchez A, Biddle JW, Steward CD, Alberti S, Bush K, Tenover FC (2008) Novel carbapenem-hydrolyzing β-lactamase KPC-1 from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother 52:809
Shen P, Wei Z, Jiang Y, Du X, Ji S, Yu Y, Li L (2009) Novel genetic environment of the carbapenem-hydrolyzing beta-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob Agents Chemother 53:4333–4338
Funding
The present study was supported in part by the Key Technologies Research and Development Program for Infectious Diseases of China (no. 2009ZX10004-104), the National Natural Science Foundation of China (no. 81071396), funding from the Shanghai Medical Key Discipline and the Science Foundation of Shanghai Jinshan (2008-3-5).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Li, G., Wei, Q., Wang, Y. et al. Novel genetic environment of the plasmid-mediated KPC-3 gene detected in Escherichia coli and Citrobacter freundii isolates from China. Eur J Clin Microbiol Infect Dis 30, 575–580 (2011). https://doi.org/10.1007/s10096-010-1124-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-010-1124-7