Abstract
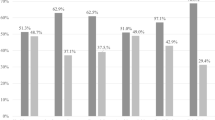
The real-time polymerase chain reaction (PCR) quantification of several vaginal bacterial groups in healthy women and patients developing asymptomatic bacterial vaginosis (BV) and candidiasis (CA) was performed. Statistical analysis revealed that the BV condition is characterised by a great variability among subjects and that it is associated with a significant increase of Prevotella, Atopobium, Veillonella and Gardnerella vaginalis, and a drop in Lactobacillus. On the contrary, the vaginal microflora of healthy women and patients developing CA was found to be homogeneous and stable over time.


Similar content being viewed by others
References
Fredricks DN, Fiedler TL, Marrazzo JM (2005) Molecular identification of bacteria associated with bacterial vaginosis. N Engl J Med 353:1899–1911 doi:10.1056/NEJMoa043802
Sobel JD, Chaim W (1996) Vaginal microbiology of women with acute recurrent vulvovaginal candidiasis. J Clin Microbiol 34:2497–2499
Rönnqvist PD, Forsgren-Brusk UB, Grahn-Håkansson EE (2006) Lactobacilli in the female genital tract in relation to other genital microbes and vaginal pH. Acta Obstet Gynecol Scand 85:726–735
Hyman RW, Fukushima M, Diamond L, Kumm J, Giudice LC, Davis RW (2005) Microbes on the human vaginal epithelium. Proc Natl Acad Sci USA 102:7952–7957 doi:10.1073/pnas.0503236102
Eschenbach DA (1993) History and review of bacterial vaginosis. Am J Obstet Gynecol 169:441–445
Thies FL, König W, König B (2007) Rapid characterization of the normal and disturbed vaginal microbiota by application of 16S rRNA gene terminal RFLP fingerprinting. J Med Microbiol 56:755–761 doi:10.1099/jmm.0.46562-0
Zdolsek B, Hellberg D, Fröman G, Nilsson S, Mårdh PA (1995) Vaginal microbiological flora and sexually transmitted diseases in women with recurrent or current vulvovaginal candidiasis. Infection 23:81–84 doi:10.1007/BF01833870
Gibbs RS (2007) Asymptomatic bacterial vaginosis: is it time to treat? Am J Obstet Gynecol 196:495–496 doi:10.1016/j.ajog.2007.04.001
Zariffard MR, Saifuddin M, Sha BE, Spear GT (2002) Detection of bacterial vaginosis-related organisms by real-time PCR for Lactobacilli, Gardnerella vaginalis and Mycoplasma hominis. FEMS Immunol Med Microbiol 34:277–281 doi:10.1111/j.1574-695X.2002.tb00634.x
Matsuki T, Watanabe K, Fujimoto J, Takada T, Tanaka R (2004) Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces. Appl Environ Microbiol 70:7220–7228 doi:10.1128/AEM.70.12.7220-7228.2004
Matsuki T, Watanabe K, Fujimoto J, Miyamoto Y, Takada T, Matsumoto K, Oyaizu H, Tanaka R (2002) Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl Environ Microbiol 68:5445–5451
Rinttilä T, Kassinen A, Malinen E, Krogius L, Palva A (2004) Development of an extensive set of 16S rDNA-targeted primers for quantification of pathogenic and indigenous bacteria in faecal samples by real-time PCR. J Appl Microbiol 97:1166–1177 doi:10.1111/j.1365-2672.2004.02409.x
Heilig HGHJ, Zoetendal EG, Vaughan EE, Marteau P, Akkermans ADL, de Vos WM (2002) Molecular diversity of Lactobacillus spp. and other lactic acid bacteria in the human intestine as determined by specific amplification of 16S ribosomal DNA. Appl Environ Microbiol 68:114–123 doi:10.1128/AEM.68.1.114-123.2002
Sha BE, Chen HY, Wang QJ, Zariffard MR, Cohen MH, Spear GT (2005) Utility of Amsel criteria, Nugent score, and quantitative PCR for Gardnerella vaginalis, Mycoplasma hominis, and Lactobacillus spp. for diagnosis of bacterial vaginosis in human immunodeficiency virus-infected women. J Clin Microbiol 43:4607–4612 doi:10.1128/JCM.43.9.4607-4612.2005
Vitali B, Pugliese C, Biagi E, Candela M, Turroni S, Bellen G, Donders GG, Brigidi P (2007) Dynamics of vaginal bacterial communities in women developing bacterial vaginosis, candidiasis, or no infection, analyzed by PCR-denaturing gradient gel electrophoresis and real-time PCR. Appl Environ Microbiol 73:5731–5741 doi:10.1128/AEM.01251-07
De Backer E, Verhelst R, Verstraelen H, Alqumber MA, Burton JP, Tagg JR, Temmerman M, Vaneechoutte M (2007) Quantitative determination by real-time PCR of four vaginal Lactobacillus species, Gardnerella vaginalis and Atopobium vaginae indicates an inverse relationship between L. gasseri and L. iners. BMC Microbiol 7:115 doi:10.1186/1471-2180-7-115
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Biagi, E., Vitali, B., Pugliese, C. et al. Quantitative variations in the vaginal bacterial population associated with asymptomatic infections: a real-time polymerase chain reaction study. Eur J Clin Microbiol Infect Dis 28, 281–285 (2009). https://doi.org/10.1007/s10096-008-0617-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10096-008-0617-0