Abstract
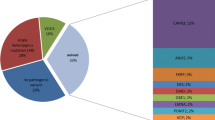
Genetic testing is being considered the first-step in the investigation of hereditary myopathies. However, the performance of the different testing approaches is little known. The aims of the present study were to evaluate the diagnostic yield of a next-generation sequencing panel comprising 39 genes as the first-tier test for genetic myopathies diagnosis and to characterize clinical and molecular findings of families from southern Brazil. Fifty-one consecutive index cases with clinical suspicion of genetic myopathies were recruited from October 2014 to March 2018 in a cross-sectional study. The overall diagnostic yield of the next-generation sequencing panel was 52.9%, increasing to 60.8% when including cases with candidate variants. Multi-gene panel solved the diagnosis of 12/25 (48%) probands with limb-girdle muscular dystrophies, of 7/14 (50%) with congenital muscular diseases, and of 7/10 (70%) with muscular dystrophy with prominent joint contractures. The most frequent diagnosis for limb-girdle muscular dystrophies were LGMD2A/LGMD-R1-calpain3-related and LGMD2B/LGMD-R2-dysferlin-related; for congenital muscular diseases, RYR1-related-disorders; and for muscular dystrophy with prominent joint contractures, Emery-Dreifuss-muscular-dystrophy-type-1 and COL6A1-related-disorders. In summary, the customized next-generation sequencing panel when applied in the initial investigation of genetic myopathies results in high diagnostic yield, likely reducing patient’s diagnostic odyssey and providing important information for genetic counseling and participation in disease-specific clinical trials.


Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Walton JN (1954) On the classification and natural history of the myopathies. Trans Am Neurol Assoc 13:19–21
Norwood FL, Harling C, Chinnery PF et al (2009) Prevalence of genetic muscle disease in Northern England: in-depth analysis of a muscleclinic population. Brain 132:3175–3186
Amburgey K, McNamara N, Bennet LR, McCormick ME, Acsadi G, Dowling JJ (2011) Prevalence of congenital myopathies in a representativepediatric United States population. Ann Neurol 70:662–665
Dimachkie MM, Barohn RJ (2014) Distal myopathies. Neurol Clin 32:817–842
Mah JK, Korngut L, Fiest KM, Dykeman J, Day LJ, Pringsheim T et al (2015) A systematic review and meta-analysis on the epidemiology of the muscular dystrophies. Can J Neurol Sci 43:163–177
Cohen BH (2019) Mitochondrial and metabolic myopathies. Continuum (Minneap Minn) 25:1732–1766
Wicklund MP (2019) The limb-girdle muscular dystrophies. Continuum (Minneap Minn) 25:1599–1618
Butterfield RJ (2019) Congenital muscular dystrophy and congenital myopathy. Continuum (Minneap Minn) 25:1640–1661
Winckler PB, Da Silva AMS, Coimbra-Neto AR, Carvalho E, Cavalcanti EBU, Sobreira CFR, Marrone CD, Machado-Costa MC, Carvalho AAS, Feio RHF, Rodrigues CL, Gonçalves MVM, Tenório RB, Mendonça RH, Cotta A, Paim JFO, Costa e Silva C, de Aquino Cruz C, Bená MI, Betancur DFA, El Husny AS, de Souza ICN, Duarte RCB, Reed UC, Chaves MLF, Zanoteli E, França MC Jr, Saute JA (2019) Clinicogenetic lessons from 370 patients with autosomal recessive limb-girdle muscular dystrophy. Clin Genet 96:341–353
Mercuri E, Bönnemann CG, Muntoni F (2019) Muscular dystrophies. Lancet 394(10213):2025–2038
Jackson CE, Barohn RJ (2013) A pattern recognition approach to myopathy. Continuum (Minneap Minn) 19:1674–97
Pasnoor M, Dimachkie MM (2019) Approach to muscle and neuromuscular junction disorders. Continuum (Minneap Minn) 25:1536–63
Wattjes MP, Kley RA, Fischer D (2010) Neuromuscular imaging in inherited muscle diseases. Eur Radiol 20:2447–2460
Pillen S, Arts IMP, Zwarts MJ (2008) Muscle ultrasound in neuromuscular disorders. Muscle Nerve 37:679–693
Nigro V, Savarese M (2016) Next-generation sequencing approaches for the diagnosis of skeletal muscle disorders. Curr Opin Neurol 29:621–627
Kress W, Rost S, Kolokotronis K, Meng G, Pluta N, Müller-Reible C (2017) The genetic approach: next-generation sequencing-based diagnosis of congenital and infantile myopathies/muscle dystrophies. Neuropediatrics 48:242–246
Magri F, Nigro V, Angelini C, Mongini T, Mora M, Moroni I et al (2017) The Italian LGMD registry: relative frequency, clinical features, and differential diagnosis. Muscle Nerve 55:55–68
Rubegni A, Malandrini A, Dosi C, Astrea G, Baldacci J, Battisti C, Bertocci G, Donati MA, Dotti MT, Federico A, Giannini F, Grosso S, Guerrini R, Lenzi S, Maioli MA, Melani F, Mercuri E, Sacchini M, Salvatore S, Siciliano G, Tolomeo D, Tonin P, Volpi N, Santorelli FM, Cassandrini D (2019) Next-generation sequencing approach to hyperCKemia: a 2-year cohort study. Neurol Genet 16(5):e352
Chakravorty S, Nallamilli BRR, Khadilkar SV, Singla MB, Bhutada A, Dastur R, Gaitonde PS, Rufibach LE, Gloster L, Hegde M (2020) Clinical and genomic evaluation of 207 genetic myopathies in the Indian subcontinent. Front Neurol 11:559327
Kopanos C, Tsiolkas V, Kouris A, Chapple CE, Albarca Aguilera M, Meyer R, Massouras A (2019) VarSome: the human genomic variant search engine. Bioinformatics 35:1978–1980
Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP (2011) Integrative genomics viewer. Nat Biotechnol 29:24–26
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, ACMG Laboratory Quality Assurance Committee (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17:405–24
Abou Tayoun AN, Pesaran T, DiStefano MT, Oza A, Rehm HL, Biesecker LG, Harrison SM, ClinGen Sequence Variant Interpretation Working Group (ClinGen SVI) (2018) Recommendations for interpreting the loss of function PVS1 ACMG/AMP variant criterion. Hum Mutat 39:1517–1524
Biesecker LG, Harrison SM, ClinGen Sequence Variant Interpretation Working Group (2018) The ACMG/AMP reputable source criteria for the interpretation of sequence variants. Genet Med 20:1687–1688
Brnich SE, Abou Tayoun AN, Couch FJ, Cutting GR, Greenblatt MS, Heinen CD, Kanavy DM, Luo X, McNulty SM, Starita LM, Tavtigian SV, Wright MW, Harrison SM, Biesecker LG, Berg JS, Clinical Genome Resource Sequence Variant Interpretation Working Group (2019) Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework. Genome Med 12:3
Adzhubei IA, Schmidt S, Peshkin L et al (2010) A method and server for predicting damaging missense mutations. Nat Methods 7:248–249
Ng PC, Henikoff S (2001) Predicting deleterious amino acid substitutions. Genome Res 11(5):863–874
Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M (2019) CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res 47:D886–D894
Jagadeesh KA, Wenger AM, Berger MJ, Guturu H, Stenson PD, Cooper DN, Bernstein JA, Bejerano G (2016) M-CAP eliminates a majority of variants of uncertain significance in clinical exomes at high sensitivity. Nat Genet 48:1581–1586
Schwarz JM, Cooper DN, Schuelke M, Seelow D (2014) MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods 11:361–362
Ioannidis NM, Rothstein JH, Pejaver V, Middha S, McDonnell SK, Baheti S, Musolf A, Li Q, Holzinger E, Karyadi D, Cannon-Albright LA, Teerlink CC, Stanford JL, Isaacs WB, Xu J, Cooney KA, Lange EM, Schleutker J, Carpten JD, Powell IJ, Cussenot O, Cancel-Tassin G, Giles GG, MacInnis RJ, Maier C, Hsieh CL, Wiklund F, Catalona WJ, Foulkes WD, Mandal D, Eeles RA, Kote-Jarai Z, Bustamante CD, Schaid DJ, Hastie T, Ostrander EA, Bailey-Wilson JE, Radivojac P, Thibodeau SN, Whittemore AS, Sieh W (2016) REVEL: an ensemble method for predicting the pathogenicity of rare missense variants. Am J Hum Genet 99:877–885
Desmet FO, Hamroun D, Lalande M, Collod-Béroud G, Claustres M, Béroud C (2009) Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 37:e67
Cooper GM, Stone EA, Asimenos G et al (2005) Distribution and intensity of constraint in mammalian genomic sequence. Genome Res 15:901–913
Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, Gauthier LD, Brand H, Solomonson M, Watts NA, Rhodes D, Singer-Berk M, England EM, Seaby EG, Kosmicki JA, Walters RK, Tashman K, Farjoun Y, Banks E, Poterba T, Wang A, Seed C, Whiffin N, Chong JX, Samocha KE, Pierce-Hoffman E, Zappala Z, O’Donnell-Luria AH, Minikel EV, Weisburd B, Lek M, Ware JS, Vittal C, Armean IM, Bergelson L, Cibulskis K, Connolly KM, Covarrubias M, Donnelly S, Ferriera S, Gabriel S, Gentry J, Gupta N, Jeandet T, Kaplan D, Llanwarne C, Munshi R, Novod S, Petrillo N, Roazen D, Ruano-Rubio V, Saltzman A, Schleicher M, Soto J, Tibbetts K, Tolonen C, Wade G, Talkowski ME, Genome Aggregation Database Consortium, Neale BM, Daly MJ, MacArthur DG (2020) The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581:434–443
Fowler A, Mahamdallie S, Ruark E, Seal S, Ramsay E, Clarke M, Uddin I, Wylie H, Strydom A, Lunter G, Rahman N (2016) Accurate clinical detection of exon copy number variants in a targeted NGS panel using DECoN. Wellcome Open Res 1:20
Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215
Savarese M, Di Fruscio G, Torella A, Fiorillo C, Magri F, Fanin M, Ruggiero L, Ricci G, Astrea G, Passamano L, Ruggieri A, Ronchi D, Tasca G, D’Amico A, Janssens S, Farina O, Mutarelli M, Marwah VS, Garofalo A, Giugliano T, Sampaolo S, Del Vecchio BF, Esposito G, Piluso G, D’Ambrosio P, Petillo R, Musumeci O, Rodolico C, Messina S, Evilä A, Hackman P, Filosto M, Di Iorio G, Siciliano G, Mora M, Maggi L, Minetti C, Sacconi S, Santoro L, Claes K, Vercelli L, Mongini T, Ricci E, Gualandi F, Tupler R, De Bleecker J, Udd B, Toscano A, Moggio M, Pegoraro E, Bertini E, Mercuri E, Angelini C, Santorelli FM, Politano L, Bruno C, Comi GP, Nigro V (2016) The genetic basis of undiagnosed muscular dystrophies and myopathies: Results from 504 patients. Neurology 87:71–76
Gonzalez-Quereda L, Rodriguez MJ, Diaz-Manera J, Alonso-Perez J, Gallardo E, Nascimento A, Ortez C, Natera-de Benito D, Olive M, Gonzalez-Mera L, Munain AL, Zulaica M, Poza JJ, Jerico I, Torne L, Riera P, Milisenda J, Sanchez A, Garrabou G, Llano I, Madruga-Garrido M, Gallano P (2020) Targeted next-generation sequencing in a large cohort of genetically undiagnosed patients with neuromuscular disorders in Spain. Genes (Basel) 11:539
Winder TL, Tan CA, Klemm S, White H, Westbrook JM, Wang JZ, Entezam A, Truty R, Nussbaum RL, McNally EM, Aradhya S (2020) Clinical utility of multigene analysis in over 25,000 patients with neuromuscular disorders. Neurol Genet 6:e412
Dardas Z, Swedan S, Al-Sheikh Qassem A, Azab B (2020) The impact of exome sequencing on the diagnostic yield of muscular dystrophies in consanguineous families. Eur J Med Genet 63:103845
Magri F, Nigro V, Angelini C, Mongini T, Mora M, Moroni I, Toscano A, D’angelo MG, Tomelleri G, Siciliano G, Ricci G, Bruno C, Corti S, Musumeci O, Tasca G, Ricci E, Monforte M, Sciacco M, Fiorillo C, Gandossini S, Minetti C, Morandi L, Savarese M, Fruscio GD, Semplicini C, Pegoraro E, Govoni A, Brusa R, Del Bo R, Ronchi D, Moggio M, Bresolin N, Comi GP (2017) The Italian limb girdle muscular dystrophy registry: relative frequency, clinical features, and differential diagnosis. Muscle Nerve 55(1):55–68. https://doi.org/10.1002/mus.25192
Nallamilli BRR, Chakravorty S, Kesari A, Tanner A, Ankala A, Schneider T, da Silva C, Beadling R, Alexander JJ, Askree SH, Whitt Z, Bean L, Collins C, Khadilkar S, Gaitonde P, Dastur R, Wicklund M, Mozaffar T, Harms M, Rufibach L, Mittal P, Hegde M (2018) Genetic landscape and novel disease mechanisms from a large LGMD cohort of 4656 patients. Ann Clin Transl Neurol 5:1574–1587
Passos-Bueno MR, Vainzof M, Moreira ES, Zatz M (1999) Seven autosomal recessive limb-girdle muscular dystrophies in the Brazilian population: from LGMD2A to LGMD2G. Am J Med Genet 82:392–398
Ghaoui R, Cooper ST, Lek M, Jones K, Corbett A, Reddel SW, Needham M, Liang C, Waddell LB, Nicholson G, O’Grady G, Kaur S, Ong R, Davis M, Sue CM, Laing NG, North KN, MacArthur DG, Clarke NF (2015) Use of whole-exome sequencing for diagnosis of limb-girdle muscular dystrophy: outcomes and lessons learned. JAMA Neurol 72:1424–1432
Schofield D, Alam K, Douglas L, Shrestha R, MacArthur DG, Davis M, Laing NG, Clarke NF, Burns J, Cooper ST, North KN, Sandaradura SA, O’Grady GL (2017) Cost-effectiveness of massively parallel sequencing for diagnosis of paediatric muscle diseases. NPJ Genom Med 2:4
François-Heude MC, Walther-Louvier U, Espil-Taris C, Beze-Beyrie P, Rivier F, Baudou E, Uro-Coste E, Rigau V, Martin Negrier ML, Rendu J, Morales RJ, Pégeot H, Thèze C, Lacourt D, Coville AC, Cossée M, Cances C (2021) Evaluating next-generation sequencing in neuromuscular diseases with neonatal respiratory distress. Eur J Paediatr Neurol 31:78–87
Wang Y, Peng W, Guo HY, Li H, Tian J, Shi YJ, Yang X, Yang Y, Zhang WQ, Liu X, Liu GN, Deng T, Sun YM, Xing WL, Cheng J, Feng ZC (2016) Next-generation sequencing-based molecular diagnosis of neonatal hypotonia in Chinese population. Sci Rep 6:29088
Heller SA, Shih R, Kalra R, Kang PB (2020) Emery-Dreifuss muscular dystrophy. Muscle Nerve 61:436–448
Zanoteli E, Soares PS, Silva AMSD, Camelo CG, Fonseca ATQSM, Albuquerque MAV, Moreno CAM, Lopes Abath Neto O, Novo Filho GM, Kulikowski LD, Reed UC (2020) Clinical features of collagen VI-related dystrophies: a large Brazilian cohort. Clin Neurol Neurosurg 192:105734
Fan Y, Liu A, Wei C, Yang H, Chang X, Wang S, Yuan Y, Bonnemann C, Wu Q, Wu X, Xiong H (2018) Genetic and clinical findings in a Chinese cohort of patients with collagen VI-related myopathies. Clin Genet 93:1159–1171
Acknowledgements
The authors are grateful to the patients and families for their participation in this study and to the professionals who attended these individuals but were not directly involved in this research project.
Funding
The study was funded by the Fundo de Incentivo à Pesquisa e Eventos-HCPA (Grant Number: 17–0340) and by unrestricted research grant from PTC Therapeutics.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval
The study was approved by the Ethics in Research Committee of our institution (GPPG-HCPA/17–0340).
Informed consent
Informed written consent was obtained from all individuals’/guardians’ prior participation.
Conflict of interest
None.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10072_2022_5934_MOESM1_ESM.xlsx
Supplementary file1 Supplemental Table 1 Clinical, genetic, neurophysiological and neuropathological description of subjects. AA, amino acid; AD, autosomal dominant; AO, age at onset; AR, autosomal recessive; AWA, age at walking aid dependency; AWC, age at wheelchair dependency; CMD; congenital muscular diseases; DD, disease duration; DM, distal myopathy; DMD, Duchenne muscular dystrophy; EMG; electromyography; F, female; Fam, family; GMWS, modified Gardner-Medwin-Walton scale. IHC, immunohistochemistry; LGMD; limb-girdle muscular dystrophy; M, male; Max, maximum; MDJC, muscular dystrophy with prominent joint contractures; MM, metabolic myopathy; MRI, magnetic resonance imaging; NA, not available; SMA, spinal muscular atrophy; UA, ultrastructural analysis; *diagnosed based on clinical findings and family molecular diagnosis.# Not detected by the next-generation sequencing panel due to low coverage and detected by whole exome sequencing (XLSX 24 KB)
Rights and permissions
About this article
Cite this article
Winckler, P.B., Chwal, B.C., Dos Santos, M.A.R. et al. Diagnostic yield of multi-gene panel for muscular dystrophies and other hereditary myopathies. Neurol Sci 43, 4473–4481 (2022). https://doi.org/10.1007/s10072-022-05934-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10072-022-05934-y