Abstract
Objective
The IKBKE has been proven to be associated with systemic lupus erythematosus (SLE) in a genome-wide association study (GWAS) conducted by our group. The objective of the recent study is to investigate the contribution of IKBKE functional variants (rs2297550) to SLE.
Methods
We detected the regulatory effect of rs2297550 on IKBKE expression by expression quantitative trait loci (eQTL) study. Then, we investigated the differences of IKBKE mRNA expression levels in peripheral blood mononuclear cells (PBMCs) between 135 SLE patients and 130 healthy controls using quantitative real-time PCR (qRT-PCR). We further analyzed the association of SLE clinical characteristics with IKBKE mRNA expression and rs2297550 polymorphisms.
Results
The results of eQTL indicated the genotype “GG” of single-nucleotide polymorphism (SNP) rs2297550 was associated with lower expression levels of IKBKE (P = 0.022) in normal controls. Compared with the healthy control group, the expression levels of IKBKE mRNA in patients with SLE were significantly decreased (P = 2.32 × 10−12). In clinical characteristics, we found that IKBKE mRNA expression levels were associated with vasculitis (P = 0.015) and increased C-reactive protein (CRP) (P = 0.021) in SLE patients.
Conclusion
In this study, we not only detected that the variant rs2297550 of IKBKE may be closely related to SLE, but also proposed functional hypotheses for the association signals.
Key Points • The rs2297550 is located in a region with transcriptional regulatory function and may regulate the expression of IKBKE via these regulatory elements. • The genotype “GG” of SNP rs2297550 was associated with lower expression levels of IKBKE. • The expression of IKBKE mRNA was decreased in SLE patients compared with healthy controls. • IKBKE contributes to the clinical characteristics of SLE. |
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Systemic lupus erythematosus (SLE) (OMIM 152700) is a systemic autoimmune disease with various complicated clinical characteristics. Over the past several decades, genome-wide association studies (GWASs) have recognized more than 100 single nucleotide polymorphisms (SNPs) closely related to SLE [1,2,3]. Our group firstly detected SNP rs2297550 of IKBKE was associated with SLE in a trans-ethnicity GWAS between Han Chinese (P = 1.52 × 10−4) and European (P = 1.59 × 10−4). We showed the results of SLE susceptible SNP rs2297550 in that study (Supplementary Table 1) [4] and further summarized allelic and genotypic frequencies of rs2297550 in different population according to 1000 Genomes Project Phase 3 (Supplementary Table 2) [5]. IKBKE encodes protein IKKε which is an inhibitor of the nuclear factor kappa-B kinase subunit epsilon gene [6]. IKKε belongs to the noncanonical IkB kinase family and has a crucial impact in adjust of the immune response [6, 7]. IKKε is a vital member of activation and transmission of NF-κB signaling pathway [8] which is one of the important factors of the disease pathogenesis of immune system [9]. Here, we analyzed and discovered an additional role of IKBKE possibly as a mediator protecting from untoward death of the cell in SLE.
We found the genotype of rs2297550 was associated with IKBKE mRNA expression levels and explored the difference of IKBKE mRNA expression levels between SLE patients and healthy controls in peripheral blood mononuclear cells (PBMCs). We then revealed the relationship between IKBKE mRNA expression and various clinical characteristics of SLE patients. It indicated that IKBKE may play a vital role in the SLE mechanism.
Methods
Patients and healthy controls
All of 135 SLE patients and 130 healthy controls were enrolled in this study (Table 1); all were enlisted from The First Affiliated Hospital of Anhui Medical University, Hefei, Anhui province, China. The patients of SLE were diagnosed by at least two experienced physicians using the revised American College of Rheumatology SLE classification criteria [10], and their clinical manifestations were documented via a full clinical check-up. The SLEDAI score [11] was performed for each patient according to the clinical findings collected at blood collection time point. The clinical characteristics we defined for each patient were based on laboratory parameters and the medical history recorded by physicians. The clinically confirmed healthy control group had no history of SLE, family history of SLE. All subjects had no other inflammatory process such as rheumatoid arthritis, Sjögren’s syndrome, and so on. Written informed consent was signed by each subject. This study got permission from the Institutional Review Board of the Anhui Medical University of China and was based on the principles of the 1964 Declaration of Helsinki and its later amendments.
Total RNA isolation
We collected blood (5 ml) from all subjects in an anticoagulation tube and then diluted by an equal volume of phosphate buffered saline (PBS). A 15ml centrifuge tube was filled with 5 ml of Ficoll-Paque Premium (GE Healthcare) and 10 ml of the 1:1 diluted blood. PBMCs were extracted by density gradient ultracentrifugation in terms of manufacturer’s requirements. Total RNA was extracted using TRIzol Reagent. In order to obtain high purity RNA, we used the NanoDrop 2000 spectrophotometer to measure samples.
qRT-PCR analysis
Extracted RNA was reversed transcription (400 ng) with the PrimeScript RT reagent Kit (Takara). The quality of complementary DNA (cDNA) was determined by qRT-PCR. We used glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression as the internal control. The primers were employed in the qRT-PCR: IKBKE primers: forward 5′-TGTCACTGGGGCTGCAGAG-3′ and reverse 5′- GTCGAAGCCCCAGCACTTG-3′. The samples were expanded for 40 cycles on a ViiA 7 Real-Time PCR system (Applied Biosystems) under the following cycles: denatured at 95 °C for 5 seconds, annealed and extended at 60 °C for 30 seconds, finally subjected to dissociation phase. The reactions were performed in a 5 μl reaction system of 10 μM primers. The IKBKE mRNA expression levels were calculated using the 2-ΔΔCt method [12].
Genotyping
Whole blood DNA was extracted using Flexi Gene DNA kits (QIAGEN, Hilden, Germany). The SNP, rs2297550, was genotyped using the Applied Biosystems 7900HT at the State Key Laboratory of Dermatology (Hefei, Anhui, China). According to the manufacturer’s introductions, we genotyped each sample with approximately 20 ng/μl of genomic DNA. The TaqMan SNP Genotyping Assay uses 5 μl reaction volumes, containing 2.5 μl 2 × Master Mix, 0.125 μl 40 × assay consisting of primer and probe, 1 μl genomic DNA and 1.375 μl RNase-Free Water.
Functional annotation and biological insights
The function and regulatory potential of rs2297550 were assessed by using HaploRegv 4.1 (http://archive.broadinstitute.org/mammals/haploreg/haploreg.php). RegulomeDB database (http://regulome.stanford.edu/) was used to identify DNA features and regulatory elements in the human genome noncoding regions obtaining rs2297550 [13, 14]. GTEx (Genotype-Tissue Expression) Analysis Release V7 (dbGaP Accession phs000424.v7.p2) [15] was used to discover the effect of rs2297550 on IKBKE mRNA expression in the skin.
Statistical analysis
We applied logistic regression to analyze the effects of Body Mass Index (BMI), genotypes and alleles on IKBKE mRNA expression. The regulation effect of rs2297550 on IKBKE mRNA expression was analyzed by one-way analysis of variance. Because IKBKE mRNA expression did not follow the normal distribution, the difference of IKBKE mRNA expression levels between SLE patients and healthy controls was analyzed by the nonparametric Mann-Whitney test. The relationship between gene expression levels and clinical characteristics was also investigated by the nonparametric Mann-Whitney test. In addition, to explore whether the clinical characteristics of SLE are related to the different genotypes or alleles of rs2297550, we used the statistical method of χ2 test or one-way analysis of variance. We used SPSS version 24.0 software for all data analysis. Figures were plotted by GraphPad Prism (version 8.01). P values (2-tailed) less than 0.05 was considered statistically significant.
Results
Association of rs2297550 with IKBKE mRNA expression and SLE clinical characteristics
We performed eQTL study to detect the effect of rs2297550 on IKBKE mRNA expression. The result indicated the genotype “GG” of SNP rs2297550 was associated with lower expression levels of IKBKE (P = 0.022) in healthy controls (Fig. 1a). However, no significant difference was identified in SLE patients (P = 0.801) (Fig. 1b). Additionally, we analyzed the association of some clinical characteristics with different genotype or allele, respectively (Supplementary Table 3). Unfortunately, we did not find that clinical characteristics related to any particular genotype or allele. We considered the limited sample size may affect the statistic power, especially once stratifying SLE patients in clinical features.
a The effect of rs2297550 on IKBKE mRNA expression levels in PBMCs from healthy controls. Of the 128 controls, 23 individuals with CC, 71 with GC, and 34 with CC were analyzed. The group with “GG” homozygous has the lowest expression level (P = 0.022). b The effect of rs2297550 on IKBKE mRNA expression levels in PBMCs from SLE patients. Of the 118 patients, 22 individuals with CC, 55 with GC, and 41 with GG were analyzed. The IKBKE mRNA expression did not significantly correlate with genotypes of rs2297550 (P = 0.801)
Regulatory effect prediction of rs2297550
The HaploReg v4.1 database predicted that rs2297550 which span the 5′ untranslated region (UTR) of IKBKE is a binding site motif. The variant rs2297550 overlapped with gene regulatory elements, i.e., gene promoters and enhancers marked by histone modifications H3K27ac, H3K4me1, and H3K4me3 in some immune cells, kidney, and skin (Table 2). RegulomeDB showed that rs2297550 had transcription factor (TF) binding and DNase sensitivity data types. These results all support the possibility that rs2297550 is located in a region with transcriptional regulation and that the expression of IKBKE can be regulated via these regulatory elements. Accordingly, we investigated how rs2297550 affects IKBKE expression in GTEx. And the result revealed that rs2297550-G was related to the decreased mRNA expression of IKBKE in skin (sun-exposed (P = 6.1 × 10−5) and not sun exposed (P = 9 × 10−6)) tissue.
IKBKE mRNA expression analysis
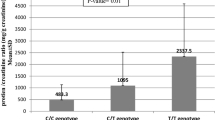
Excluding unqualified subjects, 121 SLE patients and 128 healthy controls were used for IKBKE gene expression statistics. As shown in Fig. 2, we revealed that the median expression levels of IKBKE mRNA in SLE patients (Q50 = 0.013 (Q25 = 0.009, Q75 = 0.015)) were lower than those in healthy controls (0.025 (0.014, 0.023)), with 1.92-fold increase in healthy controls compared with SLE patients (P = 2.32 × 10−12). To detect the effect from genotype and allele, we grouped SLE patients and healthy controls by genotype and allele, respectively, and then compared IKBKE mRNA expression in two groups (Supplementary Fig.1). The results were all consistent that the expression of IKBKE mRNA in SLE was decreased with respect to the controls. At the same time, we did not find that genotypes and alleles were related to the expression of IKBKE mRNA (Table 1).
The relevance of IKBKE mRNA expression with clinical characteristics
In addition, we analyzed the relevance of IKBKE mRNA expression with clinical manifestations and laboratory indicators, respectively (Table 3). We noticed that IKBKE mRNA expression levels were significantly lowered (P = 0.015) in SLE patients with vasculitis involved than those without (Fig. 3a). And it was also suggestively lowered (P = 0.021) in patients with CRP increased than those with CRP normal (Fig. 3b).
Discussion
We had reported rs2297550 located in IKBKE region is significantly associated with SLE in a published GWAS [4]. Moreover, previous studies found that the SNP rs1539241, rs12142086, and rs2151222 of IKBKE region are associated with SLE by candidate gene study [16, 17]. However, the function of these SNPs in SLE remains to be confirmed. In this study, we predicted the regulatory effect of rs2297550 by various databases, analyzed the effect of rs2297550 on IKBKE mRNA expression by eQTL, and detected the relation of IKBKE mRNA expression to disease clinical characteristics by qRT-PCR. The results provided important clues that variant rs2297550 may be involved in SLE by regulating IKBKE mRNA expression levels.
By functional annotation, we knew IKBKE 5′ UTR variant rs2297550 has both promoter and enhancer regulatory effects on certain immune cells, kidney, and skin. The rs2297550 was found to be a putative eQTL for IKBKE [4]. This study also revealed the rs2297550 can regulate the IKBKE mRNA expression level in PBMCs and the homozygous “GG” of risk allele with lower IKBKE expression levels (P = 0.022). Nevertheless, no significant differences were found in patients. We thought that the small sample size of patients, the disease itself, and the medical treatment may hide the true effect of SNP rs2297550 on IKBKE expression. In addition, we queried rs2297550 in GTEx and confirmed rs2297550-G was strongly connected with decreased mRNA expression of IKBKE in the skin. These bioinformatic insights may further support the function of rs2297550 in the regulation of IKBKE mRNA expression.
IKBKE encodes B cell kinase ε κ light chain gene enhancer inhibitor (IKKε) which participates in multiple immune signaling pathways, such as NF-κB signaling pathway [18]. The presence of IKKε would enhance the antiapoptotic function of NF-κB in response to DNA damage [19]. In addition, the lack of IKKε may active tumor necrosis factor (TNF) signaling which is strongly linked to activation NF-κB with the result of triggering the untoward death of the cell [20, 21]. Therefore, we intend to draw a role that IKBKE possibly as a mediator affect NF-κB signaling pathway and protect from untoward death of the cell in SLE. All these indicated the lower expression levels of IKBKE may contribute to the incidence and development of SLE.
Furthermore, we found that IKBKE mRNA expression levels were associated with vasculitis in SLE patients (P = 0.015). Vasculitis is characterized by inflammatory cell infiltration and subsequent vascular wall necrosis, which is one of the most characteristic processes in the clinical evolution of SLE [22]. From the above discussion, we may think that the lack of IKKε increases the untoward death of the cell and accelerates the necrosis of the vessel wall. Some studies showed that TNF stimulates the production of CRP [23,24,25]. So it is reasonable to believe that the lack of IKKε promotes TNF signaling which leads to the elevation of CRP. Therefore, in this study, the patients with lower expression levels of IKBKE tended to display higher CRP levels (P = 0.021).
In summary, we dissected the contribution of the variant rs2297550 to the susceptibility of SLE and provided particularly functional hypotheses for IKBKE that participates in the associated signals of SLE pathogenesis. Our research may contribute to improve our knowledge about the pathogenesis of SLE and provide directions for some novel drug development and clinical interventions in SLE. However, current research has some limitations. Firstly, the sample size is limited. Besides, most patients with SLE received corticosteroids or immunosuppressive treatment. These medicines may affect our results. More extensive large-scale replication and rigorous researches are necessary in the future. Larger sample size may significantly reduce the influences from medical treatment and increase the statistical power.
References
Chen L, Morris DL, Vyse TJ (2017) Genetic advances in systemic lupus erythematosus: an update. Curr Opin Rheumatol 29(5):423–433. https://doi.org/10.1097/BOR.0000000000000411
Langefeld CD, Ainsworth HC, Cunninghame Graham DS, Kelly JA, Comeau ME, Marion MC, Howard TD, Ramos PS, Croker JA, Morris DL, Sandling JK, Almlof JC, Acevedo-Vasquez EM, Alarcon GS, Babini AM, Baca V, Bengtsson AA, Berbotto GA, Bijl M, Brown EE, Brunner HI, Cardiel MH, Catoggio L, Cervera R, Cucho-Venegas JM, Dahlqvist SR, D'Alfonso S, Da Silva BM, de la Rua FI, Doria A, Edberg JC, Endreffy E, Esquivel-Valerio JA, Fortin PR, Freedman BI, Frostegard J, Garcia MA, de la Torre IG, Gilkeson GS, Gladman DD, Gunnarsson I, Guthridge JM, Huggins JL, James JA, Kallenberg CGM, Kamen DL, Karp DR, Kaufman KM, Kottyan LC, Kovacs L, Laustrup H, Lauwerys BR, Li QZ, Maradiaga-Cecena MA, Martin J, McCune JM, McWilliams DR, Merrill JT, Miranda P, Moctezuma JF, Nath SK, Niewold TB, Orozco L, Ortego-Centeno N, Petri M, Pineau CA, Pons-Estel BA, Pope J, Raj P, Ramsey-Goldman R, Reveille JD, Russell LP, Sabio JM, Aguilar-Salinas CA, Scherbarth HR, Scorza R, Seldin MF, Sjowall C, Svenungsson E, Thompson SD, Toloza SMA, Truedsson L, Tusie-Luna T, Vasconcelos C, Vila LM, Wallace DJ, Weisman MH, Wither JE, Bhangale T, Oksenberg JR, Rioux JD, Gregersen PK, Syvanen AC, Ronnblom L, Criswell LA, Jacob CO, Sivils KL, Tsao BP, Schanberg LE, Behrens TW, Silverman ED, Alarcon-Riquelme ME, Kimberly RP, Harley JB, Wakeland EK, Graham RR, Gaffney PM, Vyse TJ (2017) Transancestral mapping and genetic load in systemic lupus erythematosus. Nat Commun 8:16021. https://doi.org/10.1038/ncomms16021
Deng Y, Tsao BP (2017) Updates in lupus genetics. Curr Rheumatol Rep 19(11):68. https://doi.org/10.1007/s11926-017-0695-z
Morris DL, Sheng Y, Zhang Y, Wang YF, Zhu Z, Tombleson P, Chen L, Cunninghame Graham DS, Bentham J, Roberts AL, Chen R, Zuo X, Wang T, Wen L, Yang C, Liu L, Yang L, Li F, Huang Y, Yin X, Yang S, Ronnblom L, Furnrohr BG, Voll RE, Schett G, Costedoat-Chalumeau N, Gaffney PM, Lau YL, Zhang X, Yang W, Cui Y, Vyse TJ (2016) Genome-wide association meta-analysis in Chinese and European individuals identifies ten new loci associated with systemic lupus erythematosus. Nat Genet 48(8):940–946. https://doi.org/10.1038/ng.3603
Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, GA MV, Abecasis GR (2015) A global reference for human genetic variation. Nature 526(7571):68–74. https://doi.org/10.1038/nature15393
Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T (2003) IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol 4(5):491–496. https://doi.org/10.1038/ni921
Hacker H, Karin M (2006) Regulation and function of IKK and IKK-related kinases. Sci STKE 2006(357):re13. https://doi.org/10.1126/stke.3572006re13
Bonizzi G, Karin M (2004) The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol 25(6):280–288. https://doi.org/10.1016/j.it.2004.03.008
Roman-Blas JA, Jimenez SA (2006) NF-kappaB as a potential therapeutic target in osteoarthritis and rheumatoid arthritis. Osteoarthr Cartil 14(9):839–848. https://doi.org/10.1016/j.joca.2006.04.008
Hochberg MC (1997) Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum 40(9):1725. https://doi.org/10.1002/1529-0131(199709)40:9<1725::AID-ART29>3.0.CO;2-Y
Bombardier C, Gladman DD, Urowitz MB, Caron D, Chang CH (1992) Derivation of the SLEDAI. A disease activity index for lupus patients. The Committee on Prognosis Studies in SLE. Arthritis Rheum 35(6):630–640. https://doi.org/10.1002/art.1780350606
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Ward LD, Kellis M (2012) HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res 40(Database issue):D930–D934. https://doi.org/10.1093/nar/gkr917
Demirci FY, Wang X, Kelly JA, Morris DL, Barmada MM, Feingold E, Kao AH, Sivils KL, Bernatsky S, Pineau C, Clarke AE, Ramsey-Goldman R, Vyse TJ, Gaffney PM, Manzi S, Kamboh MI (2016) Identification of a new susceptibility locus for systemic lupus erythematosus on chromosome 12 in individuals of European ancestry. Arthritis Rheumatol 68(1):174–183. https://doi.org/10.1002/art.39403
Gibson G (2015) Human genetics. GTEx detects genetic effects. Science 348(6235):640–641. https://doi.org/10.1126/science.aab3002
Wang C, Ahlford A, Laxman N, Nordmark G, Eloranta ML, Gunnarsson I, Svenungsson E, Padyukov L, Sturfelt G, Jonsen A, Bengtsson AA, Truedsson L, Rantapaa-Dahlqvist S, Sjowall C, Sandling JK, Ronnblom L, Syvanen AC (2013) Contribution of IKBKE and IFIH1 gene variants to SLE susceptibility. Genes Immun 14(4):217–222. https://doi.org/10.1038/gene.2013.9
Sandling JK, Garnier S, Sigurdsson S, Wang C, Nordmark G, Gunnarsson I, Svenungsson E, Padyukov L, Sturfelt G, Jonsen A, Bengtsson AA, Truedsson L, Eriksson C, Rantapaa-Dahlqvist S, Malarstig A, Strawbridge RJ, Hamsten A, Criswell LA, Graham RR, Behrens TW, Eloranta ML, Alm G, Ronnblom L, Syvanen AC (2011) A candidate gene study of the type I interferon pathway implicates IKBKE and IL8 as risk loci for SLE. Eur J Hum Genet 19(4):479–484. https://doi.org/10.1038/ejhg.2010.197
Maher P (2008) Proteasome inhibitors prevent oxidative stress-induced nerve cell death by a novel mechanism. Biochem Pharmacol 75(10):1994–2006. https://doi.org/10.1016/j.bcp.2008.02.008
Renner F, Moreno R, Schmitz ML (2010) SUMOylation-dependent localization of IKKepsilon in PML nuclear bodies is essential for protection against DNA-damage-triggered cell death. Mol Cell 37(4):503–515. https://doi.org/10.1016/j.molcel.2010.01.018
Lafont E, Draber P, Rieser E, Reichert M, Kupka S, de Miguel D, Draberova H, von Massenhausen A, Bhamra A, Henderson S, Wojdyla K, Chalk A, Surinova S, Linkermann A, Walczak H (2018) TBK1 and IKKepsilon prevent TNF-induced cell death by RIPK1 phosphorylation. Nat Cell Biol 20(12):1389–1399. https://doi.org/10.1038/s41556-018-0229-6
Zampieri S, Alaibac M, Iaccarino L, Rondinone R, Ghirardello A, Sarzi-Puttini P, Peserico A, Doria A (2006) Tumour necrosis factor alpha is expressed in refractory skin lesions from patients with subacute cutaneous lupus erythematosus. Ann Rheum Dis 65(4):545–548. https://doi.org/10.1136/ard.2005.039362
Barile-Fabris L, Hernandez-Cabrera MF, Barragan-Garfias JA (2014) Vasculitis in systemic lupus erythematosus. Curr Rheumatol Rep 16(9):440. https://doi.org/10.1007/s11926-014-0440-9
Volanakis JE (2001) Human C-reactive protein: expression, structure, and function. Mol Immunol 38(2–3):189–197. https://doi.org/10.1016/s0161-5890(01)00042-6
Collerton J, Martin-Ruiz C, Davies K, Hilkens CM, Isaacs J, Kolenda C, Parker C, Dunn M, Catt M, Jagger C, von Zglinicki T, Kirkwood TB (2012) Frailty and the role of inflammation, immunosenescence and cellular ageing in the very old: cross-sectional findings from the Newcastle 85+ study. Mech Ageing Dev 133(6):456–466. https://doi.org/10.1016/j.mad.2012.05.005
Marta W, Michal C, Grazyna N (2019) TNF-alpha G-308A genetic variants, serum CRP-hs concentration and DNA damage in obese women. Mol Biol Rep 47:855–866. https://doi.org/10.1007/s11033-019-04764-0
Acknowledgments
We thank all the study participants and all the volunteers who have so willingly participated in this study, thus making this study possible. We also appreciate the Genotype-Tissue Expression (GTEx) Project which was supported by the Common Fund of the Office of the Director of the National Institutes of Health and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS.
Funding
This article was made open access with the financial support of King’s College London. This study was supported financially by the National Key Laboratory Breeding Base of Dermatology of China (4601007103), the Key Laboratory of Dermatology Ministry of Education of China (4601007102), and the Cultivation of fund winners of innovative research groups (4601011224).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This study got permission from the Institutional Review Board of the Anhui Medical University of China and was based on the principles of the 1964 Declaration of Helsinki and its later amendment.
Disclosures
None.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhu, T., Hong, J., Shuai, Z. et al. The decreased expression of IKBKE in systemic lupus erythematosus. Clin Rheumatol 39, 2611–2617 (2020). https://doi.org/10.1007/s10067-020-05006-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10067-020-05006-6