Abstract
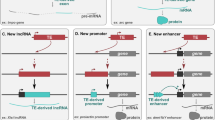
The CHRM3 gene is a member of the muscarinic acetylcholine receptor family that plays important roles in the regulation of fundamental physiological functions. The evolutionary mechanism of exon-acquisition and alternative splicing of the CHRM3 gene in relation to transposable elements (TEs) were analyzed using experimental approaches and in silico analysis. Five different transcript variants (T1, T2, T3, T3-1, and T4) derived from three distinct promoter regions (T1: L1HS, T2, T4: original, T3, T3-1: THE1C) were identified. A placenta (T1) and testis (T3 and T3-1)-dominated expression pattern appeared to be controlled by different TEs (L1HS and THE1C) that were integrated into the common ancestor genome during primate evolution. Remarkably, the T1 transcript was formed by the integration event of the human specific L1HS element. Among the 12 different brain regions, the brain stem, olfactory region, and cerebellum showed decreased expression patterns. Evolutionary analysis of splicing sites and alternative splicing suggested that the exon-acquisition event was determined by a selection and conservation mechanism. Furthermore, continuous integration events of transposable elements could produce lineage specific alternative transcripts by providing novel promoters and splicing sites. Taken together, exon-acquisition and alternative splicing events of CHRM3 genes were shown to have occurred through the continuous integration of transposable elements following conservation.
Similar content being viewed by others
References
Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSIBLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402.
Bièche, I., Laurent, A., Laurendeau, I., Duret, L., Giovangrandi, Y., Frendo, J.L., Olivi, M., Fausser, J.L., Evain-Brion, D., and Vidaud, M. (2003). Placenta-specific INSL4 expression is mediated by a human endogenous retrovirus element. Biol. Reprod. 68, 1422–1429.
Braverman, A.S., Tallarida, R.J., and Ruggieri, M.R. (2008). The use of occupation isoboles for analysis of a response mediated by two receptors: M2 and M3 muscarinic receptor subtype-induced mouse stomach contractions. J. Pharmacol. Exp. Ther. 325, 954–960.
Brett, D., Hanke, J., Lehmann, G., Haase, S., Delbrück, S., Krueger, S., Reich, J., and Bork, P. (2000). EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett. 474, 83–86.
Calarco, J.A., Xing, Y., Caceres, M., Calarco, J.P., Xiao, X., Pan, Q., Lee, C., Preuss, T.M., and Blencowe, B.J. (2007). Global analysis of alternative splicing differences between humans and chimpanzees. Genes Dev. 21, 2963–2975.
Cammeron, J.R., Loh, E.Y., and Davis, R.W. (1979). Evidence for transposition of dispersed repetitive DNA families in yeast. Cell 16, 739–751.
Dunn, C.A., and Mager, D.L. (2005). Transcription of the human and rodent SPAM1/PH-20 genes initiates within an ancient endogenous retrovirus. BMC Genomics 6, 47.
Dunn, C.A., Medstrand, P., and Mager, D.L. (2003). An endogenous retroviral long terminal repeat is the dominant promoter for human beta1,3-galactosyltransferase 5 in the colon. Proc. Natl. Acad. Sci. USA 100, 12841–12846.
Dunn, C.A., Romanish, M.T., Gutierrez, L.E., van de Lagemaat, L.N., and Mager, D.L. (2006). Transcription of two human genes from a bidirectional endogenous retrovirus promoter. Gene 266, 335–342.
Forsythe, S.M., Kogut, P.C., McConville, J.F., Fu, Y., McCauley, J.A., Halayko, A.J., Liu, H.W., Kao, A., Fernandes, D.J., Bellam, S., et al. (2002). Structure and transcription of the human m3 muscarinic receptor gene. Am. J. Respir. Cell Mol. Biol. 26, 298–305.
Gautam, D., Duttaroy, A., Cui, Y., Han, S.J., Deng, C., Seeger, T., Alzheimer, C., and Wess, J. (2006). M1-M3 muscarinic acetylcholine receptor-deficient mice: novel phenotypes. J. Mol. Neurosci. 30, 157–160.
Gerthoffer, W.T. (2005). Signal-transduction pathways that regulate visceral smooth muscle function. III. Coupling of muscarinic receptors to signaling kinases and effector proteins in gastrointestinal smooth muscles. Am. J. Physiol. Gastrointest. Liver Physiol. 288, G849–853.
Grindley, N.D.F., and Reed, R.R. (1985). Transpositional recombination in prokaryotes. Ann. Rev. Biochem. 54, 863–896.
Guo, Y., Traurig, M., Ma, L., Kobes, S., Harper, I., Infante, A.M., Bogardus, C., Baier, L.J., and Prochazka, M. (2006). CHRM3 gene variation is associated with decreased acute insulin secretion and increased risk for early-onset type 2 diabetes in Pima Indians. Diabetes 55, 3625–3629.
Huh, J.W., Ha, H.S., Kim, D.S., and Kim, H.S. (2008). Placentarestricted expression of LTR-derived NOS3. Placenta 29, 602–608.
International Human Genome Sequencing Consortium (2001). Initial sequencing and analysis of the human genome. Nature 409, 860–921.
Jurka, J. (2000). Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 16, 418–420.
Kim, D.S., Kim, T.H., Huh, J.W., Kim, I.C., Kim, S.W., Park, H.S., and Kim, H.S. (2006). LINE FUSION GENES: a database of LINE expression in human genes. BMC Genomics 7, 139.
Kitazawa, T., Hashiba, K., Cao, J., Unno, T., Komori, S., Yamada, M., Wess, J., and Taneike, T. (2007). Functional roles of muscarinic M2 and M3 receptors in mouse stomach motility: studies with muscarinic receptor knockout mice. Eur. J. Pharmacol. 554, 212–222.
Landry, J.R., and Mager, D.L. (2003). Functional analysis of the endogenous retroviral promoter of the human endothelin B receptor gene. J. Virol. 77, 7459–7466.
Lev-Maor, G., Sorek, R., Shomron, N., and Ast, G. (2003). The birth of an alternatively spliced exon: 3’ splice-site selection in Alu exons. Science 300, 1288–1291.
Mätlik, K., Redik, K., and Speek, M. (2006). L1 antisense promoter drives tissue-specific transcription of human genes. J. Biomed. Biotechnol. 1, 71753.
Matsui, M., Araki, Y., Karasawa, H., Matsubara, N., Taketo, M.M., and Seldin, M.F. (1999). Mapping of five subtype genes for muscarinic acetylcholine receptor to mouse chromosomes. Genes Genet. Syst. 74, 15–21.
McClintock, B. (1956). Controlling elements and the gene. Cold Spring Harbor Symp. Quant. Biol. 35, 243–251.
Mills, R.E., Bennett, E.A., Iskow, R.C., Luttig, C.T., Tsui, C., Pittard, W.S., and Devine, S.E. (2006). Recently mobilized transposons in the human and chimpanzee genomes. Am. J. Hum. Genet. 78, 671–679.
Mills, R.E., Bennett, E.A., Iskow, R.C., and Devine, S.E. (2007). Which transposable elements are active in the human genome? Trends Genet. 23, 183–191.
Renuka, T.R., Ani, D.V., and Paulose, C.S. (2004). Alterations in the muscarinic M1 and M3 receptor gene expression in the brain stem during pancreatic regeneration and insulin secretion in weanling rats. Life Sci. 75, 2269–2280.
Rio, D.C. (1990). Molecular mechanisms regulating Drosophila P element transposition. Ann. Rev. Gen. 24, 543–578.
Sela, N., Mersch, B., Gal-Mark, N., Lev-Maor, G., Hotz-Wagenblatt, A., and Ast, G. (2007). Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu’s unique role in shaping the human transcriptome. Genome Biol. 8, R127.
Speek, M. (2001). Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol. Cell. Biol. 21, 1973–1985.
Sverdlov, E.D. (2000). Retroviruses and primate evolution. Bioessays 22, 161–171.
Urrutia, A.O., Ocaña, L.B., and Hurst, L.D. (2008) Do Alu repeats drive the evolution of the primate transcriptome? Genome Biol. 9, R25.
van de Lagemaat, L.N., Landry, J.R., Mager, D.L., and Medstrand, P. (2003). Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet. 19, 530–536.
Wall, S.J., Yasuda, R.P., Li, M., and Wolfe, B.B. (1991). Development of an antiserum against m3 muscarinic receptors: distribution of m3 receptors in rat tissues and clonal cell lines. Mol. Pharmacol. 40, 783–789.
Watanabe, H., Fujiyama, A., Hattori, M., Taylor, T.D., Toyoda, A., Kuroki, Y., Noguchi, H., BenKahla, A., Lehrach, H., Sudbrak, R., et al. (2004). DNA sequence and comparative analysis of chimpanzee chromosome 22. Nature 429, 382–328.
Yamada, M., Miyakawa, T., Duttaroy, A., Yamanaka, A., Moriguchi, T., Makita, R., Ogawa, M., Chou, C.J., Xia, B., Crawley, J.N., et al. (2001). Mice lacking the M3 muscarinic acetylcholine receptor are hypophagic and lean. Nature 410, 207–212.
Zhang, W., Yamada, M., Gomeza, J., Basile, A.S., and Wess, J. (2002). Multiple muscarinic acetylcholine receptor subtypes modulate striatal dopamine release, as studied with M1-M5 muscarinic receptor knock-out mice. J. Neurosci. 22, 6347–6352.
Zhang, H.M., Chen, S.R., Matsui, M., Gautam, D., Wess, J., and Pan, H.L. (2006). Opposing functions of spinal M2, M3, and M4 receptor subtypes in regulation of GABAergic inputs to dorsal horn neurons revealed by muscarinic receptor knockout mice. Mol. Pharmacol. 69, 1048–1055.
Zhang, H.M., Zhou, H.Y., Chen, S.R., Gautam, D., Wess, J., and Pan, H.L. (2007). Control of glycinergic input to spinal dorsal horn neurons by distinct muscarinic receptor subtypes revealed using knockout mice. J. Pharmacol. Exp. Ther. 232, 963–971.
Author information
Authors and Affiliations
Corresponding author
Additional information
These authors contributed equally to this work.
About this article
Cite this article
Huh, JW., Kim, YH., Lee, SR. et al. Gain of new exons and promoters by lineage-specific transposable elements-integration and conservation event on CHRM3 gene. Mol Cells 28, 111–117 (2009). https://doi.org/10.1007/s10059-009-0106-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10059-009-0106-z