Abstract
CHROMSCAN implements a composite likelihood model for the analysis of association data. Disease-gene localisation is on a linkage disequilibrium unit (LDU) map, and locations and standard errors, for putatively causal polymorphisms, are determined by the programme. Distortions of the probability distribution created by auto-correlation are avoided by implementation of a permutation test. We evaluated the relative efficiency of the LDU map by simulating pseudo-phenotypes in real genotype samples. We observed that multi-locus mapping on an underlying LDU map reduces location error by ∼46%. Furthermore, there is a small, but significant, increase in power of ∼5%. Effective meta-analysis across multiple samples, increasingly important to combine evidence from genome-wide and other association data, is achieved through the weighted combination of location evidence provided by the programme.
Similar content being viewed by others
Introduction
Many genome-wide association mapping studies, using high-density panels of single nucleotide polymorphisms (SNPs), are underway. Typically, these involve hundreds to thousands of DNA samples of cases and controls and are screened for several hundred thousand SNPs across the genome. Efficient analysis and interpretation of these vast data sets raises many difficulties. If significance is tested at individual SNPs, huge numbers of false positive results are generated, making interpretation difficult. Sub-optimal statistical adjustments for the number of tests may under-correct or lose power. However, models that utilise information simultaneously from multiple SNPs offer advantages by reducing the overall number of tests and recovering additional information. Reliable computation of significance levels remains an issue given the large numbers of non-independent SNPs and consequent auto-correlation. Other concerns recognise the difficulty of incorporating information on the underlying linkage disequilibrium (LD) structure when testing association with phenotype. This is important because using this information has been shown to substantially increase power and precision for mapping (Maniatis et al. 2004, 2005).
We describe and evaluate here the CHROMSCAN programme, which models association with disease at multiple SNPs in regions defined by non-overlapping sliding windows. The programme rapidly identifies regions for follow-up and determines reliable P values, maximum likelihood locations and standard errors for putatively causal polymorphisms. CHROMSCAN employs a composite-likelihood-based model, estimating a small number of parameters in each region, thereby greatly reducing the number of tests made (Collins and Morton 1998). A permutation test ensures the P value distribution is not distorted through the auto-correlation created by extensive LD (Morton et al. 2007). Association with disease is modelled on a map in LD units (LDUs) (Maniatis et al. 2002; Lau et al. 2007), which is an LD analogue of the genetic linkage map. We also developed and describe an efficient parallel computing version that greatly accelerates the analysis of high-density genome-scan data. This programme, CHROMSCAN-cluster, runs on a Linux-based computing cluster.
We examined the resolution of this approach for localising pseudo-phenotypes simulated in a real SNP genotype sample and analysed in regions of at least ten LD units. Simulation in a real sample retains the LD structure, variations in marker density, marker informativeness and other peculiarities encountered in real genotype samples. We dichotomised randomly selected SNPs to produce a pseudo-phenotype and predicted the location from analysis of the remaining neighbouring SNPs to compare the resolution and relative power of the LDU map to the corresponding kilobase (kb) map. Distances between the known and predicted locations were evaluated in both LDUs and kb.
Methods
Core methodology
CHROMSCAN implements a modification of the Malecot model, representing association, z, between SNPs and disease on an underlying marker map: \(z_i=(1-L) {Me^{-\epsilon |S_{i}-S|}} +L,\) where S i is the location of the ith SNP in LDU (alternatively kb) and the S parameter represents the map location showing maximal association with disease (Morton et al. 2007). The ε parameter describes the decline of association with map distance, M is the intercept and the asymptote is estimated (L) or predicted (L p). The predicted asymptote is defined (Morton et al. 2001; Zhang et al. 2002) as the mean absolute value of a standard normal deviate, weighted by the information K z = n(a + b)(b + d)/(a + c)(cc + d), where a, b, , d are counts from the 2 × 2 table of alleles by affection status, totalling n haplotypes. However, association with disease is typically evaluated with SNP diplotype data, and therefore, the 3 × 2 table of genotype counts by affection status is reduced to the corresponding 2 × 2 table, scoring n haplotypes from n/2 diplotypes (Maniatis 2007, Table 1). Counts a, b, c, d, are assigned such that a and b represent SNP alleles coded “1” and “2”, respectively, in the controls and c and d the corresponding counts in cases, arranged so ad–bc ≥ 0, b ≤ c. For association with disease at each marker: \( \hat z\, = \,(ad - bc)/(a + b)(b + d), \) and \( \chi ^2 _1 \, = \,\hat z^2 \user2{K}{_z}, \) where \( 0\, < \,\user2{K}_z \, = \,\chi ^2 _1 /\hat z^2 \le n. \) The composite likelihood is lk = e−Λ/2, where \(\Uplambda = \sum {_i \user2{K}_{zi} (\hat z_i - z_i )^2.} \) Model fitting employs the dfpmin function from Press et al. (1992), and convergence to the global maximum is achieved by beginning iteration from a large number of points in the parameter space. Two models are contrasted in the programme: a “flat” model, which assumes no association with disease (model “A”) taking L = L p and M = 0, and an association model (“D”), which estimates M, S and L. For both models, the values Λ are computed from the contrast with the baseline model: L = M = 0. As the baseline and model A do not test association with disease, there is no location estimate, S. CHROMSCAN obtains the difference X = ΛA − ΛD for the real data (H 1) and a large number of replicates (H 0), as X j for the jth replicate, for which the phenotype is randomised (shuffled). Shuffling creates data sets under the null that are consistent with, although not equivalent to, model A. P values under H 0, (P j), are computed from fractional ranks in the sample of replicates. From P j the corresponding χ2 j3 is computed from the NAG g01fcc function (http://www.nag.co.uk/) and hence the variance V j as: X j/χ2 j3. Variances for the replicates, V j, under H 0, are used to predict the variance V under H 1. Computation of the corresponding variance for H 1 requires a sorted sub-set of replicates, centred as far as possible on X, and the model: ln V j = A + B ln X j. Where feasible, X is centred between the 20 closest replicates with X j ≤ X and the corresponding 20 with X j ≥ X; if X is an outlier, the 20 closest values are taken. V under H 1 is estimated as exp(A + B ln X), giving χ2 3 = X/V (Morton et al. 2007), with corresponding probability from the g01ecc NAG sub-routine. Estimation of V from replicates under H 0 therefore avoids distortion due to auto-correlation and enables the computation of χ2 3 (H 1) and the standard error of location S. Simultaneous estimates of M, S and L provide an information matrix that is inverted, and the nominal variance K SS is obtained. Then the information K about S is computed as: (1/K SS)/(V/3) with the standard error \( \sqrt {1/K}. \)
Programme options
CHROMSCAN accepts genotype (=diplotype) or phase-known haplotype data in fixed format files, with columns representing SNPs and rows unrelated individuals. Labelling of SNP alleles is “1” or “2”, with missing coded as dot or blank, affected individuals coded “1” and controls “0”. The data file specifies column locations for each SNP and sequence locations in LDU and kb from the p-telomere or arbitrary offset. CHROMSCAN accepts a candidate region, delimited by two kb locations, or scans a chromosome in non-overlapping regions. A minimum of 30 SNPs is assigned to each region, and remaining SNPs at the end of the map are assigned to the final region. The width in LDU of regions has a default of at least 10 LDUs. The populations represented in HapMap (http://www.hapmap.org) have 58–81,000 LDUs (Lau et al. 2007), so a high-density genome-wide scan yields ∼6–8,000 regions for analysis.
Optionally, CHROMSCAN determines locations on the kb map, allowing comparisons of power and precision with the LDU map. The ε parameter, for which 1/ε is the “swept radius” (the average extent of LD that is useful for mapping), is ∼1 for LDU maps and ∼0.02 for kb maps (corresponding to ∼50 kb, Morton et al. 2007). These estimates are obtained from the LDMAP programme: (http://www.som.soton.ac.uk/research/geneticsdiv/epidemiology/LDMAP/default.htm).
SNPs are screened in the control sample for deviations from Hardy–Weinberg equilibrium and an optional χ2 cutoff imposed. This enables identification and removal of SNPs with distorted genotype distributions that may reflect technical difficulties in genotyping, or other sources of error, but does not guarantee that the remaining markers are in Hardy–Weinberg equilibrium.
Finally, the number of replicates for each region in which the phenotype is randomised is specified. A relatively small number of replicates (for example, 1,000) for the first pass through genome-wide data efficiently determines a sub-set of nominally significant regions for more thorough analysis. For a sub-set of regions strongly associated with the phenotype, >10,000 replicates may be required to give precise P values.
CHROMSCAN-cluster parallel version
CHROMSCAN-cluster is a wrapper programme encapsulating CHROMSCAN and is implemented on a Linux Beowulf cluster. Batch queuing and job management is administrated by Open-PBS (Portable Batch System), http://www.openpbs.org/. The programme divides consecutive regions into batches and submits these in parallel. The number of regions per batch is user definable for adjusting the batch load according to SNP density. The programme features synchronous processing supporting multiple SNP data set submissions. To efficiently utilise dual-processor machines in the cluster, batches are assigned as two jobs per submission. In addition to job monitoring commands (i.e. “showq” and “qstat”), supplied by Open-PBS, a custom-made programme “checkStatus” tracks the status of the submitted jobs grouped by SNP data set. Modification of the software for local systems should be straightforward, as the source is written in standard C. A parallel version of CHROMSCAN for a Linux cluster is available from: http://www.som.soton.ac.uk/research/geneticsdiv/epidemiology/chromscan/#sourceCode
Simulations comparing utility of LDU and kb maps
Maniatis et al. (2004) described simulations in which single SNPs were selected from a sample of genotypes and the allelic count (0, 1, or 2, for the three genotypes) was used as a pseudo-phenotype, the location of which was predicted from the other markers as a test of mapping resolution and power. We adapted this approach for a case-control study using genotypes described by Klein et al. (2005). The authors undertook a genome-wide association study with 116,204 markers and 96 cases with age-related macular degeneration and 50 controls. We discarded all of the disease phenotypic data and used the genotypes obtained for chromosome 4 to construct pseudo-phenotypes with known location. The SNP genotypic data, with markers approximately every 26 kb across the genome, are at the lower end of current densities in ongoing genome-wide studies (typically with at least 500,000 SNPs, http://www.wtccc.org.uk/info/overview.shtml, Wellcome Trust Case Control Consortium 2007). Low marker density data presents a challenge for predicting the location of the pseudo-phenotype from neighbouring markers and enables a useful comparison of the utility of alternative underlying maps. For constructing the pseudo-phenotype, we selected SNPs spaced roughly evenly across the chromosome and dichotomised the SNP genotype. We assigned individuals with missing genotypes or any heterozygotes (1, 2 genotypes) alternately as “case” or “control” to yield approximately equal numbers of both. Individuals with the homozygote genotypes 1, 1 or 2, 2 were coded as “case” and “control”, respectively. Using this pseudo-phenotype, the location of the selected SNP was then predicted using only association with the remaining SNPs in the region. We rejected a proportion of the samples generated, analysing only those yielding ∼7.8 ≤ χ2 3 ≤ ∼21.1 (0.05 ≥ P ≥ 0.0001), for the A–D model comparison, to approximate the significance levels obtained in some candidate regions.
For marker locations, we used the HapMap Phase-II-derived LDU map of chromosome 4 described by Lau et al. (2007) for the CEPH (CEU) population and available from: (http://www.som.soton.ac.uk/research/geneticsdiv/epidemiology/LDMAP/map2.htm).
The LDU locations for any markers in the sample that are not given in the map were obtained by interpolation from their known kb locations and flanking markers with both kb and LDU locations. At total of 6,547 SNPs span 190,733.41 kb or 3,518.29 LDUs, after removal of 1,937 SNPs with minor allele frequencies <0.05, which were not included in any analyses. This gave an average of one SNP every 29 kb for chromosome 4. A total of 10,000 replicates were generated for each region sufficient to ensure that P values were computed with high precision across all samples.
We analysed regions across chromosome 4 that satisfied the power criteria (supplementary Table 1), permitting comparisons of the relative utility of LDU and kb maps and yielding information about the limits of mapping resolution in relatively low marker-density samples.
Results
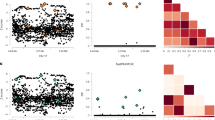
A total of 26 data sets were simulated, each with a SNP selected and dichotomised to represent a case-control phenotype (pseudo-phenotype). The LDU map of the region around one of the SNPs (rs2048070) is shown in Fig. 1, (see also supplementary Table 1). This region contains 29 SNPs (after removal of the SNP selected to define the pseudo-phenotype) spanning 579.4 kb and 22.0 LDUs. The true location of the selected SNP (not used in the model fitting) was 10,865.24 kb/373.22 LDU. The estimated location was at 10,879.22 kb/373.60 LDU, a distance of 13.98 kb/0.38 LDU from the known location.
Linkage disequilibrium unit (LDU) map of the rs2048070 region. The LDU map indicates a recombination intense region around 10,700 kb and the relatively high LD region within which the rs2048070 SNP (selected to form a pseudo-phenotype) is located (shown by the triangle). The predicted location from the fitted model is shown by the circle a few kilobases from the known location
The mean location error across the 26 data sets, for the LDU map, is 0.42 LDUs, with a standard deviation (SD) of 0.31 (Table 1). The minimum error is 0.0 and the maximum is 0.96. All of the pseudo-phenotypes are therefore mapped to well within one LDU of the correct location and the majority to within ∼0.5 LDUs. LDU locations may be converted by interpolation into approximate kb locations. Interpolation uses the nearest SNPs in the map, which flank the target LDU location, and their known kb and LDU locations to estimate the kb location that corresponds to the target LDU position. However, conversion is subject to some error when the target location lies within a block over which LDU = 0 and the kb location at the mid-point of the block is taken. When the LDU locations are interpolated, the mean error is 33.3 kb (SD 31.6), indicating very high mapping resolution. Furthermore, in higher marker-density genome scans, the resolution is likely to increase markedly. Localisation on an underlying kb rather than an LDU map is less efficient, and the mean location error increases by ∼47% to 48.8 kb. The difference in location errors between interpolated LDU and kb maps is significant at P = 0.035.
The relative power, as measured by χ2 3 for the A–D comparison (see Methods), is higher for the LDU map compared with the kb map (mean χ2 3 13.6 vs. 12.9). The difference is significant at P = 0.041 in the paired samples test and corresponds to an average increase in χ2 3 of 5% when using the LDU map (Table 1). The results for both mapping resolution and power are broadly consistent with the simulation study described by Maniatis et al. (2004), which included samples with much higher power, and the single real-data example with very high power described by Maniatis et al. (2005).
Discussion
The CHROMSCAN programme analyses genome-wide and candidate-region association data to determine reliable significance levels and efficiently reduces the impact of multiple testing through model fitting. The latter has the additional advantage of increasing power and exploiting information on the underlying LD structure in the form of an LDU map. The underlying LDU map reduces location error for mapping by almost 50% and provides significantly increased power. Implementation for parallel processing on a cluster enables rapid computation of stable P values from large numbers of replicates.
Recent genome-wide association studies have employed multi-stage designs where, following an initial genome-wide scan, increasingly large samples are tested to confirm significance. This provides a relatively economical and practical strategy and undoubtedly yields a proportion of the causal variants. However, the relatively small samples used in the initial genome-wide scan, incomplete coverage of the genome in the scan, acceptance in the first stage of relatively modest significance levels and a small number of regions for follow-up contributes to the strong possibility that important variants are overlooked. Therefore, the combination of evidence from genome scans through meta-analysis may provide additional regions worthy of further study while adding value to studies already undertaken.
Amongst possible approaches to meta-analysis is the Genome Search Meta-analysis (GSMA) method (Wise et al. 1999), which has been developed for genome-wide linkage studies. For linkage, the authors advocate the use of 30-cM bins within which the evidence for linkage is assessed and bins are ranked according to the strength of evidence. The statistic testing for linkage is formed from the sum of ranks, and significance can be evaluated from a distribution or by simulation (Levinson et al. 2003). This useful strategy could be adapted to examine genome-wide association data, perhaps in bins of 10 LDUs as we used in this study. Each bin would span approximately 500 kb/0.5 cM and would therefore provide much finer resolution than the 30-cM linkage screen. However, ranking of bins formed in this way neglects the within-bin location information and information weights produced by CHROMSCAN. An alternative simple approach to meta-analysis, which avoids bins (Morton et al. 2007), assumes s independent samples, of which the ith contributes a probability P i that is uniformly distributed on the null hypothesis. Then −2 ln P i is distributed as \( \chi _{\text{2}}^{\text{2}}, \) with \( \chi _{{\text{2}}s}^{\text{2}} = - 2\sum \ln P_i. \) Both this and GSMA approaches are applicable to data lacking a location estimate S i and information K i but have the disadvantages of assuming equal weights for samples with different information and not providing a point estimate, which would be expected to become more accurate as sample sizes increase. An alternative meta-analysis approach (Morton et al. 2007) computes a weighted location from s independent samples, assuming the same LDU map is used for all samples, as: \( \bar S\, = \,\sum_{i = 1}^s {S_i K_i /\sum {K_i } }. \) Weighting locations by information computed in CHROMSCAN provides an appropriate χ2 test and test of heterogeneity in the meta-analysis. This method has the advantage that accessions of data, in the form of additional genome scans, would be expected to reduce further target regions of interest whilst increasing significance for any putatively causal variants. However, the impact of variation between samples, including differences in allele frequencies and ascertainment, and other sources of heterogeneity must be considered in the determination of the operating characteristics of this and other approaches to meta-analysis. It seems likely that there will be increasing opportunities to apply meta-analysis with the release of genome-wide data sets on application by researchers. These include data from the Wellcome Trust Case Control Consortium (WTCCC, http://www.wtccc.org.uk/, Wellcome Trust Case Control Consortium 2007) and cancer samples from the Cancer Genetic Markers of Susceptibility study (http://cgems.cancer.gov/).
We have developed an approach, implemented in the programme CHROMSCAN for genome-wide association data analysis, which provides robustly determined P values and high power and mapping precision through the use of an underlying LDU map. Analysis within regions defined on that map suggests a framework for future meta-analyses using a GSMA-type approach, although a potentially more informative method would combine across studies the location and information evidence produced by the programme.
References
Collins A, Morton NE (1998) Mapping a disease locus by allelic association. Proc Natl Acad Sci USA 95:1741–1745
Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J (2005) Complement factor H polymorphism in age-related macular degeneration. Science 308(5720):385–389
Lau W, Kuo TY, Tapper W, Cox S, Collins A (2007) Exploiting large scale computing to construct high resolution linkage disequilibrium maps of the human genome. Bioinformatics 23(4):517–519
Levinson DF, Levinson MD, Segurado R, Lewis CM (2003) Genome scan meta-analysis of schizophrenia and bipolar disorder, Part I: methods and power analysis. Am J Hum Genet 73:17–33
Maniatis N (2007) Linkage disequilibrium maps and disease association mapping. In: Collins AR (ed) Linkage disequilibrium and association mapping: analysis and applications. Humana Press, Totowa
Maniatis N, Collins A, Xu C-F, McCarthy LC, Hewett DR, Tapper W, Ennis S, Ke X, Morton NE (2002) The first linkage disequilibrium (LD) maps: delineation of hot and cold blocks by diplotype analysis. Proc Natl Acad Sci USA 99(4):2228–2233
Maniatis N, Collins A, Gibson J, Zhang W, Tapper W, Morton NE (2004) Positional cloning by linkage disequilibrium. Am J Hum Genet 74(5):846–855
Maniatis N, Morton NE, Gibson J, Xu C-F, Hosking LK, Collins A (2005) The optimal measure of linkage disequilibrium reduces error in association mapping of affection status. Hum Mol Genet 14(1):145–153
Morton NE, Zhang W, Taillon-Miller P, Ennis S, Kwok P-Y, Collins A (2001) The optimal measure of allelic association. Proc Natl Acad Sci USA 98(9):5217–5221
Morton N, Maniatis N, Zhang W, Ennis S, Collins A (2007) Genome scanning by composite likelihood. Am J Hum Genet 80(1):19–28
Press WH, Teukolsky SA, Vetterling WT, Flannery BP (1992) Numerical recipes in C. Cambridge University Press, Cambridge
Wellcome Trust Case Control Consortium (2007) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447(7145):661–678
Wise LH, Lanchbury JS, Lewis CM (1999) Meta-analysis of genome searches. Ann Hum Genet 63:263–272
Zhang W, Collins A, Maniatis N, Tapper W, Morton NE (2002) Properties of linkage disequilibrium (LD) maps. Proc Natl Acad Sci USA 99:17004–17007
Acknowledgments
This research is supported by a University of Southampton e-Science Centre Postgraduate Research grant. The authors thank Josephine Hoh and colleagues at Yale for the genotypes used in the simulation study.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Collins, A., Lau, W. CHROMSCAN: genome-wide association using a linkage disequilibrium map. J Hum Genet 53, 121–126 (2008). https://doi.org/10.1007/s10038-007-0226-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-007-0226-2
Keywords
This article is cited by
-
Linkage disequilibrium maps for European and African populations constructed from whole genome sequence data
Scientific Data (2019)
-
Genome-wide association of breast cancer: composite likelihood with imputed genotypes
European Journal of Human Genetics (2011)
-
Composite likelihood-based meta-analysis of breast cancer association studies
Journal of Human Genetics (2011)
-
Allelic Association: Linkage Disequilibrium Structure and Gene Mapping
Molecular Biotechnology (2009)