Abstract
A total of 264 unrelated breast/ovarian cancer patients and 45 healthy individuals with familial antecedents referred for genetic testing were scanned for germ-line mutations in BRCA1 and BRCA2 by conformation-sensitive gel electrophoresis (CSGE) and heteroduplex analysis by capillary array electrophoresis (HA-CAE). We detected 101 distinct mutations (41 in BRCA1 and 60 in BRCA2); ten of them have not been previously reported. These mutations were c.2411_2429dup19, c.2802_2805delCAAA and c.5294A>G (p.E1725E) of BRCA1; and c.667C>T (p.Q147X), c.2683C>T (p.Q819X), c.5344_5347delAATA, c.5578_5579delAA;insT, c.8260_8261insGA, c.744+14C>T and c.8099A>G (p.Y2624C) of BRCA2. Twenty-four different mutations, including seven of the new mutations (five frameshift and two nonsense), were classified as pathogenic. These 24 alterations were found in 39 families (12.6% of all families). A remarkable proportion of deleterious mutations were found in BRCA2: 25 families carried a mutation in BRCA2 (BRCA2+; 64.1%) compared with 14 families BRCA1+ (35.9%). The highest incidences of deleterious mutations were found in families with three or more cases of site-specific breast cancer (BC) (27.4%) and families with BC and ovarian cancer (22.2%). Finally, four recurrent mutations, 3036_3039delACAA, c.5374_5377delTATG of BRCA2, as well as c.5272-1G>A and c.5242C>A (p.A1708E) of BRCA1, accounted for 44% of all of the deleterious mutations.
Similar content being viewed by others
Introduction
Breast cancer (BC) is the most common malignancy among women in industrialized countries. It is estimated that 5–10% of all BCs are due to genetic predisposition. Mutations in the tumor-suppressor genes BRCA1 (MIM#113705) and BRCA2 (MIM#600185) confer an increased risk of developing BC and ovarian cancer (OC) (Ford et al. 1998). However, mutations in both genes only explain approximately 20% of families with evidence of inherited susceptibility to breast cancer (Wooster and Weber 2003). Disease-causing mutations are distributed throughout the entire coding regions of both genes (The BIC database: http://www.research.nhgri.nih.gov/bic/Member/index.shtml). Apart from specific ethnic groups, there are no predominant mutations accounting for the majority of inherited breast cancer cases, and deleterious mutations present significant ethnic and geographic variation. Since the identification of BRCA1 and BRCA2 as the principal genes responsible for inherited breast cancer (Miki et al. 1994; Tavtigian et al. 1996), more than 3,400 distinct DNA sequence variants have been described in the BIC database, of which 1,723 can be classified as pathogenic, including 1,422 truncating mutations (1,064 frameshift and 358 nonsense) and 301 splicing alterations. The remaining mutations, including 1,346 distinct missense changes, are unclassified variants of uncertain clinical consequences.
With a view to offering genetic counseling to BC and OC patients from Castilla y Leon (Spain), we have scanned the entire coding regions and the exon–intron boundaries of BRCA1 and BRCA2. In previous reports we showed that our BC/OC patients had a wide range of mutations (Velasco-Sampedro et al. 2002; Duran et al. 2003; Diez et al. 2003). In fact, we had already identified a total of 27 novel DNA changes, illustrating the high diversity of BRCA1/2 mutations in our population (Velasco-Sampedro et al. 2002; Duran et al. 2003; Velasco et al. 2005).
Here we present the mutation-detection data corresponding to a set of 309 families analyzed by conformation-sensitive gel electrophoresis (CSGE) and heteroduplex analysis with capillary array electrophoresis (HA-CAE), the new high-throughput method developed by our group (Esteban-Cardenosa et al. 2004). As result of this work, we describe for the first time seven deleterious mutations (two of BRCA1 and five of BRCA2), one BRCA2 missense mutation, and two polymorphic variants (one in each BRCA gene).
Patients and methods
Patients
Ethylenediaminetetraacetic acid (EDTA)-anticoagulated blood samples as well as written informed consent were received from 264 unrelated BC/OC patients and 45 healthy individuals with familial antecedents. BC/OC patients were classified into groups based on the following criteria:
-
I.
One BC before the age of 40 (67 families)
-
II.
Two BCs in the same family before the age of 50 (33 families)
-
III.
Three or more cases of BC in the same family (62 families)
-
IV.
BC and OC in the same family or patient (27 families)
-
V.
One male BC (14 families)
-
VI.
Bilateral BC (30 families)
-
VII.
Two OCs (no families)
-
VIII.
One BC plus familial aggregation of other types of cancer (four families)
Additionally, 27 unrelated BC patients of unknown familial antecedents also participated (no relatives with BC or OC were available). Finally, 125 relatives of individuals with deleterious mutations or unclassified variants were also studied. DNA from all samples was extracted using the QIAampDNA Blood mini kit (Qiagen, Hilden, Germany).
BRCA1 and BRCA2 mutational analysis
Seventy-three PCR fragments (31 for BRCA1 and 42 for BRCA2) were amplified in 23 multiplex PCR groups. Thermocycling conditions were performed in a GeneAmp PCR system 9700 as described before (Esteban-Cardenosa et al. 2004). The mutation-detection method was either standard CSGE (92 families) (Ganguly et al. 1993; Cebrian et al. 2002) or fluorescent HA-CAE (217 families) in an ABI3100 DNA sequencer (Applied Biosystems) (Esteban-Cardenosa et al. 2004). In this case, one primer of each pair of the PCR reactions was fluorescently labeled with FAM, HEX or NED.
DNA sequencing
Fragments showing a CSGE- or HA-CAE-altered pattern were sequenced with the BigDye Terminator Sequencing Kit v3.1 (Applied Biosystems) with unlabeled forward and reverse primers.
Mutation nomenclature
Nucleotide positions were numbered considering the cDNA reference sequences U14680 (BRCA1) and U43746 (BRCA2). Description of mutations was according to accepted nomenclature guidelines of the Human Genome Variation Society (http://www.genomic.unimelb.edu.au/mdi/mutnomen/).
Results and discussion
Novel mutations
The mutational analysis of BRCA1 and BRCA2 by CSGE and HA-CAE (Fig. 1) revealed a total of 101 different mutations, 41 in BRCA1 and 60 in BRCA2, in 309 families (Tables 1 and 2). Ten novel mutations were characterized, including five frameshift (two deletions, one insertion, one duplication and one combined deletion/insertion mutation) and two nonsense mutations (Table 1), as well as one missense, one synonymous and one intronic variant mutation (Table 2). These new mutations were c.2802-2805delCAAA, c.2411-2429dup19 and c.5294 A>G (p.E1725E) of BRCA1; and c.667C>T (p.Q147X), c.2683C>T (p.Q819X), c.5344-5347delAATA, c.5578-5579delAA;insT, c.8260_8261insGA, c.744+14C>T and c.8099A>G (p.Y2624C) of BRCA2. Together with our previous results (Velasco-Sampedro et al. 2002; Duran et al. 2003; Diez et al. 2003; Velasco et al. 2005), novel mutations represent 29% (37 out of 128 different DNA changes detected in 459 unrelated families) of all mutations identified, thus illustrating the high variability of the BRCA genes.
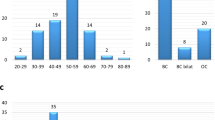
Examples of heteroduplex analysis by capillary array electrophoresis (HA-CAE) patterns. Novel pathogenic mutations c.5344_5347delAATA from BRCA2 exon fragment 11R labeled with FAM and c.2683C>T (p.Q819X) from BRCA2 exon fragment 11D labeled with NED are shown (Esteban-Cardenosa et al. 2004). Sizes of DNA standard Genescan 500-ROX are indicated
Pathological mutations
Twenty-four different disease-causing mutations (9 in BRCA1 and 15 in BRCA2) were identified in 39 unrelated families (12.6% of all the families), including seven of the novel mutations, which create premature stop codons, truncating the BRCA proteins (Table 1). Taking into account the previous description of another four novel truncating mutations (Duran et al. 2003), a total of 11 different new pathogenic mutations have been exclusively identified in our patients, indicating that each population will show a specific mutational spectrum. The two new frameshift mutations of BRCA1, c.2411_2429dup19 and c.2802_2805delCAAA, took place in exon 11. Both were detected in families with more than three BC cases (group III). With regard to BRCA2, the novel two nonsense mutations, p.Q147X and p.Q819X, and three frameshift mutations, c.5344_5347delAATA (two families), c.5578_5579delAA;insT and c.8260_8261ins GA, were identified in exons 5, 11 and 18. These five mutations were found in six unrelated patients of families of criteria groups I, III (two families), IV (two families), and one healthy individual with two familial antecedents with BC (c.8260_8261ins GA).
BRCA1/BRCA2 ratio of pathogenic mutations
We have found 14 unrelated BRCA1+ families (35.9%) and 25 BRCA2+ families (64.1%). When taking into account our previous results (Velasco-Sampedro et al. 2002; Duran et al. 2003; Diez et al. 2003), the final BRCA1/BRCA2 ratio did not change, with 23 and 42 deleterious mutations in BRCA1 (35.4%) and BRCA2 (64.6%), respectively (Table 1). A previous study of the Spanish Breast Cancer Consortium, which comprised families from our first five selection criteria (Diez et al. 2003), did not show significant differences between BRCA1 and BRCA2 (53% BRCA1+ versus 47% BRCA2+).
Apart from a putative specific feature of our population, there are several alternative reasons for this BRCA1/BRCA2 imbalance. First, a majority of BRCA2 deleterious mutations (one BRCA1+ versus six BRCA2+) were reported in young Spanish BC women from the Mediterranean area (Martinez-Ferrandis et al. 2003). Our data support these results, but only three pathogenic mutations have been found in group I.
One additional explanation may be an underrepresentation of families with BC and OC, group IV (only 8.7% of our families), where mutations in BRCA1 are more frequent than in BRCA2 (Risch et al. 2001). Moreover, a higher incidence of BRCA2 mutations has been previously reported in Japanese site-specific BC families (our group III) (Ikeda et al. 2001). In fact, we have found 5 BRCA1+ families and 12 BRCA2+ families in group III.
Another cause may be a high frequency of BRCA1 rearrangements, which are undetectable by traditional PCR-based mutation detection methods such as CSGE and SSCP. However, this possibility has been recently excluded, as only three patients carried this sort of mutations (data not shown).
Finally, another possibility is the absence of the most prevalent mutation of BRCA1 in Spain, c.187_188delAG of Ashkenazi Jews (Diez et al. 1998, 2003) and the abundance of recurrent mutations of BRCA2 in our population. This effect has been previously reported in Iceland, where a single BRCA2 frameshift mutation, c.999del5, accounts for 40% of male BCs and 7% of female BCs (Tulinius et al. 2002). The most prevalent mutation was c.3036_3039delACAA of BRCA2, which was found in seven families in this study. In addition, it was detected in other four families in a preceding report (Duran et al. 2003), making a total of 11 unrelated families. This 4-bp deletion is also the most frequent BRCA2 mutation in Spain (Diez et al. 2003; Salazar et al. 2005), and it has been detected in many European countries (Neuhausen et al. 1998). Other important recurrent mutations of BRCA2 were c.5374_5377delTATG, c.6126delT and c.9538_9539delAA (Table 1). These mutations represent more than one-third of all pathogenic mutations and may be founder mutations in our population, although this point has to be corroborated. Probably, the combination of all these hypotheses may explain the BRCA1/BRCA2 imbalance of our BC/OC patients.
The most frequent BRCA1 mutations were c.5272-1G>A and the missense mutation c.5242C>A (p.A1708E, Table 1) whose pathogenicity has been previously demonstrated (Vallon-Christersson et al. 2001). These two alterations are responsible for 39% of all deleterious mutations of BRCA1. Concerning the mutation c.5272-1G>A, it was found five times in our complete cohort of BC patients, but it has also been detected in one sporadic BC case without known familial history, who was diagnosed at 54 years of age (data not shown). This DNA change is relatively rare in the BIC database (only three records in Western European and Spanish populations). Therefore, these data also suggest a putative founder effect in Spain, which has not been demonstrated yet. Finally, the third most common mutation of BRCA1 (three unrelated families, Table 1) is the splicing mutation c.330A>G (r.310_331del22) of Galician origin (Vega et al. 2001), which was detected only once in this cohort of 309 families.
Frequency of mutations by selection criteria
The distribution of deleterious mutations according to the selection criteria was 3/67 (4.5%) in group I, 3/33 (9.1%) in group II, 17/62 (27.4%) in group III, 6/27 (22.2%) in group IV, 1/14 in group V, 3/30 (10%) in group VI, 2/27 (7.4%) in BC patients with unknown familial history and 4/45 in healthy individuals with familial antecedents with BC. No mutations were found in groups VII (two ovarian cancers) and VIII (BC + familial aggregation). As expected, a great proportion of mutations were identified in patients from groups III and IV (59% of all mutations), as they showed the highest ratio of cancer-prone mutations (27.4 and 22.2%, respectively). This frequency of mutations by clinical group is similar to the results previously reported in the Spanish population (Diez et al. 2003).
Unclassified variants and polymorphisms
The vast majority of mutations, 77 variants (32 of BRCA1 and 45 of BRCA2, Table 2), could not be classified as pathogenic mutations. As previously reported in other populations (Katagiri et al. 1998), missense variants outweighed deleterious mutations (34 versus 24). Fifty-nine DNA changes were considered as mere polymorphisms, including the two novel variants c.5294A>G of BRCA1 and c.744+14C>T of BRCA2 (Table 2). The remaining 18 mutations (17 missense mutations and one intronic variant) were variants of unknown physiological effect or unclassified variants, including the novel missense mutation c.8099A>G (p.Y2624C), which affected a conserved residue of BRCA2 (Table 2). Unfortunately, familial segregation could not be performed as this patient belongs to the group of young patients without known familial antecedents. Although a great number of the 1,346 different missense mutations (one in four codons of the BRCA genes has a missense mutation) may presumably be related to the disease, they should be regarded as unclassified variants since a functional assay of the BRCA proteins is still not available. Therefore, the role of these unclassified missense mutations in BC and OC remains to be elucidated. Despite a comprehensive analysis of BRCA1 and BRCA2, there will be high-risk BC families without deleterious mutations in these genes. In fact, the cumulative evidence from several studies suggests the putative influence of additional moderate or high penetrance genes (Nathanson and Weber 2001). Their characterization will contribute to clarifying the spectrum of genes involved in familial BC.
References
Cebrian A, Ruiz-Llorente S, Cascon A, Osorio A, Martinez-Delgado B, Benitez J, Robledo M (2002) A rapid and easy method for multiple endocrine neoplasia type 1 mutation detection using conformation-sensitive gel electrophoresis. J Hum Genet 47:190–195
Diez O, Domenech M, Alonso MC, Brunet J, Sanz J, Cortes J, del Rio E, Baiget M (1998) Identification of the 185delAG BRCA1 mutation in a Spanish Gypsy population. Hum Genet 103:707–708
Diez O, Osorio A, Duran M, Martinez-Ferrandis JI, de la Hoya M, Salazar R, Vega A, Campos B, Rodriguez-Lopez R, Velasco E, Chaves J, Diaz-Rubio E, Jesus Cruz J, Torres M, Esteban E, Cervantes A, Alonso C, San Roman JM, Gonzalez-Sarmiento R, Miner C, Carracedo A, Eugenia Armengod M, Caldes T, Benitez J, Baiget M (2003) Analysis of BRCA1 and BRCA2 genes in Spanish breast/ovarian cancer patients: a high proportion of mutations unique to Spain and evidence of founder effects. Hum Mutat 22:301–312
Duran M, Esteban-Cardenosa E, Velasco E, Infante M, Miner C (2003) Mutational analysis of BRCA2 in Spanish breast cancer patients from Castilla-Leon: identification of four novel truncating mutations. Hum Mutat 21:448
Esteban-Cardenosa E, Duran M, Infante M, Velasco E, Miner C (2004) High-throughput mutation detection method to scan BRCA1 and BRCA2 based on heteroduplex analysis by capillary array electrophoresis. Clin Chem 50:313–320
Ford D, Easton DF, Stratton M, Narod S, Goldgar D, Devilee P, Bishop DT, Weber B, Lenoir G, Chang-Claude J, Sobol H, Teare MD, Struewing J, Arason A, Scherneck S, Peto J, Rebbeck TR, Tonin P, Neuhausen S, Barkardottir R, Eyfjord J, Lynch H, Ponder BA, Gayther SA, Zelada-Hedman M, Breast Cancer Linkage Consortium (1998) Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. Am J Hum Genet 62:676–689
Ganguly A, Rock MJ, Prockop DJ (1993) Conformation-sensitive gel electrophoresis for rapid detection of single-base differences in double-stranded PCR products and DNA fragments: evidence for solvent-induced bends in DNA heteroduplexes. Proc Natl Acad Sci USA 90:10325–10329
Ikeda N, Miyoshi Y, Yoneda K, Shiba E, Sekihara Y, Kinoshita M, Noguchi S (2001) Frequency of BRCA1 and BRCA2 germline mutations in Japanese breast cancer families. Int J Cancer 91:83–88
Katagiri T, Kasumi F, Yoshimoto M, Nomizu T, Asaishi K, Abe R, Tsuchiya A, Sugano M, Takai S, Yoneda M, Fukutomi T, Nanba K, Makita M, Okazaki H, Hirata K, Okazaki M, Furutsuma Y, Morishita Y, Iino Y, Karino T, Ayabe H, Hara S, Kajiwara T, Houga S, Shimizu T, Toda M, Yamazaki Y, Uchida T, Kunitomo K, Sonoo H, Kurebayashi J, Shimotsuma K, Nakamura Y, Miki Y (1998) High proportion of missense mutations of the BRCA1 and BRCA2 genes in Japanese breast cancer families. J Hum Genet 43:42–48
Martinez-Ferrandis JI, Vega A, Chirivella I, Marin-Garcia P, Insa A, Lluch A, Carracedo A, Chaves FJ, Garcia-Conde J, Cervantes A, Armengod ME (2003) Mutational analysis of BRCA1 and BRCA2 in Mediterranean Spanish women with early-onset breast cancer: identification of three novel pathogenic mutations. Hum Mutat 22:417–418
Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W et al. (1994) A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science 266:66–71
Nathanson KL, Weber BL (2001) ‘Other’ breast cancer susceptibility genes: searching for more Holy Grail. Hum Mol Genet 10:715–720
Neuhausen SL, Godwin AK, Gershoni-Baruch R, Schubert E, Garber J, Stoppa-Lyonnet D, Olah E, Csokay B, Serova O, Lalloo F, Osorio A, Stratton M, Offit K, Boyd J, Caligo MA, Scott RJ, Schofield A, Teugels E, Schwab M, Cannon-Albright L, Bishop T, Easton D, Benitez J, King MC, Ponder BAJ, Weber B, Devilee P, Borg A, Narod SA, Goldgar D (1998) Haplotype and phenotype analysis of nine recurrent BRCA2 mutations in 111 families: results of an international study. Am J Hum Genet 62:1381–1388
Osorio A, Robledo M, Albertos J, Diez O, Alonso C, Baiget M, Benitez J (1998) Molecular analysis of the six most recurrent mutations in the BRCA1 gene in 87 Spanish breast/ovarian cancer families. Cancer Lett 123:153–158
Risch HA, McLaughlin JR, Cole DEC, Rosen B, Bradley L, Kwan E, Jack E, Vesprini DJ, Kuperstein G, Abrahamson JLA, Fan I, Wong B, Narod SA (2001) Prevalence and penetrance of germline BRCA1 and BRCA2 mutations in a population series of 649 women with ovarian cancer. Am J Hum Genet 68:700–710
Salazar R, Cruz-Hernandez JJ, Sanchez-Valdivieso E, Rodriguez CA, Gomez-Bernal A, Barco E, Fonseca E, Portugal T, Gonzalez-Sarmiento R (2006) BRCA1-2 mutations in breast cancer: identification of nine new variants of BRCA1-2 genes in a population from central Western Spain. Cancer Lett 233(1):172–177
Tavtigian SV, Simard J, Rommens J, Couch F, Shattuck-Eidens D, Neuhausen S, Merajver S, Thorlacius S, Offit K, Stoppa-Lyonnet D, Belanger C, Bell R, Berry S, Bogden R, Chen Q, Davis T, Dumont M, Frye C, Hattier T, Jammulapati S, Janecki T, Jiang P, Kehrer R, Leblanc JF, Goldgar DE et al. (1996) The complete BRCA2 gene and mutations in chromosome 13q-linked kindreds. Nat Genet 12:333–337
Tulinius H, Olafsdottir GH, Sigvaldason H, Arason A, Barkardottir RB, Egilsson V, Ogmundsdottir HM, Tryggvadottir L, Gudlaugsdottir S, Eyfjord JE (2002) The effect of a single BRCA2 mutation on cancer in Iceland. J Med Genet 39:457–462
Vallon-Christersson J, Cayanan C, Haraldsson K, Loman N, Bergthorsson JT, Brondum-Nielsen K, Gerdes AM, Moller P, Kristoffersson U, Olsson H, Borg A, Monteiro AN (2001) Functional analysis of BRCA1 C-terminal missense mutations identified in breast and ovarian cancer families. Hum Mol Genet 10:353–360
Vega A, Campos B, Bressac-de-Paillerets B, Bond PM, Janin N, Douglas FS, Domenech M, Baena M, Pericay C, Alonso C, Carracedo A, Baiget M, Diez O (2001) The R71G BRCA1 is a founder Spanish mutation and leads to aberrant splicing of the transcript. Hum Mutat 17:520–521
Velasco E, Infante M, Duran M, Esteban-Cardenosa E, Lastra E, Garcia-Giron C, Miner C (2005) Rapid mutation detection in complex genes by heteroduplex analysis with capillary array electrophoresis. Electrophoresis 26:2539–2552
Velasco-Sampedro E, Esteban-Cardenosa E, Infante-Sanz M, Durán-Dominguez M, Lastra-Aras E, Garcia-Giron C, Miner-Pino C (2002) Molecular study of the BRCA1 and BRCA2 genes in 153 breast cancer families from Castilla y Leon (Spain): new nine unclassified variants identified. Med Clin (Barc) 119:441–445
Wooster R, Weber BL (2003) Breast and ovarian cancer. N Engl J Med 348:2339–2347
Acknowledgements
This work has been supported by the Junta de Castilla y León through the regional Breast Cancer Prevention Program. M. Infante and E. Esteban-Cardeñosa were recipients of fellowships from the “Fundación Burgos para la Investigación en Salud.” We are also grateful to the patients and the clinicians, especially Dr. García Girón and Dr. Lastra (Hospital General Yagüe, Burgos, Spain), who collaborated in this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Infante, M., Durán, M., Esteban-Cardeñosa, E. et al. High proportion of novel mutations of BRCA1 and BRCA2 in breast/ovarian cancer patients from Castilla-León (central Spain). J Hum Genet 51, 611–617 (2006). https://doi.org/10.1007/s10038-006-0404-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-006-0404-7
Keywords
This article is cited by
-
Contribution of BRCA1 and BRCA2 germline mutations to early onset breast cancer: a series from north of Morocco
BMC Cancer (2020)
-
Hereditary breast and ovarian cancer in Andalusian families: a genetic population study
BMC Cancer (2018)
-
Molecular characterization and clinical interpretation of BRCA1/BRCA2 variants in families from Murcia (south-eastern Spain) with hereditary breast and ovarian cancer: clinical–pathological features in BRCA carriers and non-carriers
Familial Cancer (2017)
-
Novel and recurrent BRCA1/BRCA2 mutations in early onset and familial breast and ovarian cancer detected in the Program of Genetic Counseling in Cancer of Valencian Community (eastern Spain). Relationship of family phenotypes with mutation prevalence
Familial Cancer (2013)
-
Comprehensive splicing functional analysis of DNA variants of the BRCA2 gene by hybrid minigenes
Breast Cancer Research (2012)