Abstract
Leber hereditary optic neuropathy (LHON) is characterized by the acute or subacute bilateral painless loss of central vision, predominantly in young males. G11778A is the most common mitochondrial DNA mutation responsible for the disease. Thirty-seven percent of our LHON pedigrees (which is a much higher prevalence than that generally found) carried heteroplasmic G11778A. Analyses of four large Thai LHON pedigrees spanning four to six generations strongly suggested that the transmission of the heteroplasmic G11778A mutation is under selective pressure in favour of the mutated allele and that heteroplasmy influences the disease expression.
Similar content being viewed by others
Introduction
Leber hereditary optic neuropathy (LHON) is a maternally transmitted disease characterized by the acute or subacute loss of central vision as a result of optic nerve atrophy. It occurs predominantly in males, usually in the second to third decade of life (Nikoskelainen et al. 1987; Riordan-Eva et al. 1995). Approximately 95% of all LHON families worldwide carry one of three mitochondrial DNA (mtDNA) mutations – in the ND1 gene at nucleotide position 3460, in the ND4 gene at position 11778 or in the ND6 gene at position 14484, with the 11778 mutation being the most prevalent in almost all of the world's populations (Mackey et al. 1996; Man et al. 2002), including that of Thailand (Chuenkongkaew et al. 2001; Phasukkijwatana et al. 2006). In most LHON pedigrees, these mutations are homoplasmic (every mtDNA contains the mutant allele). However, heteroplasmy (a mixture of both normal and mutant alleles) exists in about 15% of all LHON pedigrees (Chinnery et al. 2001; Newman et al. 1991; Smith et al. 1993), most likely reflecting a recent mutational event dating back to only a few generations (Savontaus 1995). It is still inconclusive as to whether the transmission of heteroplasmic LHON mutations from one generation to the next is a random or a selective process. While the study of a number of pedigrees has found a trend towards an increasing proportion of the LHON mutation in successive generations (Bolhuis et al. 1990; Howell et al. 1994; Jacobi et al. 2001; Savontaus 1995; Smith et al. 1993), suggesting a selection for mutant alleles, other studies have found inconsistent results. Consequently, currently accepted opinion questions the selective process, supporting instead a random process (Chinnery et al. 2001; Ghosh et al. 1996; Harding et al. 1995).
Approximately 50% of males and 10% of females harbouring the LHON mutation develop optic atrophy, however, the penetrance in different pedigrees appears to vary widely (Mackey et al. 1996; Man et al. 2002), and low-penetrance branching has been proposed (Howell and Mackey 1998).
Despite extensive knowledge of the clinical and mitochondrial genetic features of LHON, some characteristics of LHON pedigrees, such as the biological phenomenon of low-penetrance branching (Howell and Mackey 1998) and selective or random transmission of a heteroplasmic LHON mutation, require additional supportive empirical data. In an attempt to provide some of the data necessary to clarify the above complexities and ambiguities of this mitochondrial disease, we analysed our extensive LHON pedigrees with the G11778A mutation in a Thai genetic background.
Materials and methods
Subjects and sample collection
Four extensive pedigrees were accessed through four unrelated probands who came to the Department of Ophthalmology, Faculty of Medicine Siriraj Hospital, Bangkok, Thailand, complaining of acute blurring of vision. Complete eye examinations were performed by the ophthalmologists, and the clinical presentations were suggestive of LHON. Blood samples were then sent to our laboratory for mtDNA analysis with informed consent. History taking for date of birth, age of onset and visual symptoms was performed as detailed in Phasukkijwatana et al. (2006). Eye examinations for visual acuities (Snellen chart), colour vision (Ishihara colour plates) as well as direct ophthalmoscopic examination were carried out, and the diagnosis of LHON was performed by a neuro-ophthalmologist.
MtDNA analysis
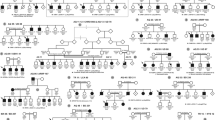
The PCR-restriction fragment length polymorphism (RFLP) analytical technique (Lertrit et al. (1999) was used to test all blood samples for the primary G11778A LHON mutation. The degree of heteroplasmy of the G11778A mutation was quantitated using radioactive restriction analysis and densitometry as described by Phasukkijwatana et al. (2006). The hypervariable segment 1 (HVS-1) in the mtDNA D-loop (nt 16024 to nt 16383) from the proband of each family was also sequenced. The entire mitochondrial genome from both the homoplasmic and the heteroplasmic branches of two pedigrees, F24 (II-2, III-22, in Fig. 1b) and F28 (IV-56, VI-39 in Fig. 1c), was sequenced. The total mitochondrial genome was also sequenced in three individuals (III-18, III-33 and IV-39, in Fig. 1d) from three branches of F30.
Four Thai LHON families with the G11778A mutation: F09 (a), F24 (b), F28 (c) and F30 (d). The pedigree symbols are as follows:









Statistical analysis
The difference in the average percentage of heteroplasmy (of each generation) between three generations of each family was tested by one-way analysis of variance (ANOVA). The Mann-Whitney test was adopted to determine the difference in average final visual acuities [latest logMAR, which is equal to log (1/Snellen visual acuity)] between heteroplasmic and homoplasmic patients. These analyses were performed using SPSS software ver. 13.0 for Windows (SPSS, Chicago, Ill.). The Kaplan-Meier survival estimates between the probability of Leber-free survival and age at onset were performed using the software Intercooled Stata 9.1 for Windows. The difference between three groups – group 1, homoplasmy (carrying the G11778A mutation load greater than 95%); group 2, carrying the G11778A mutation load between 75 and 95%; group 3, carrying the G11778A mutation load less than 75% – was compared using the log-rank test for equality of survivor functions which is also in Intercooled Stata ver. 9.1 for Windows.
Results and discussion
The pedigrees for the four unrelated LHON families with the G11778A mutation reported here consisted of four to six generations (Fig. 1). In each pedigree, mtDNA was analysed in three to five generations. Each of these LHON Thai families lived in different parts of Thailand: in the central part (F09; Fig. 1a, in Chachoengsao province), in the northeastern part (F24; Fig. 1b, in Roi Et province and F30; Fig. 1d, in Buriram province) and in the northern part (F28; Fig. 1c, in Nan province). Three of these families contained several people with the heteroplasmic mutation (F09, F24, F28), while the other family was homoplasmic (F30). All pedigrees belong to macro-haplogroup M (Tharaphan et al. 2006).
Heteroplasmic transmission
In this study we took advantage of the three large heteroplasmic pedigrees to study the transmission pattern of the heteroplamic G11778A LHON mutation. The prevalence of heteroplasmic LHON pedigrees with the G11778A mutation has been shown to be very high in Thailand – 37% (11 in 30 pedigrees) (Phasukkijwatana et al. 2006). This is similar to that found in a recent epidemiological study in the North East of England (Man et al. 2003), but higher than that mostly reported in the literature (approx. 15%) (Huoponen 2001; Man et al. 2002). If heteroplasmy does actually reflect a recent mutational event (Savontaus 1995), the recent high incidence of the G11778A mutation in Thailand presents an intriguing problem. In two of the pedigrees in our collection the G11778A mutation appeared to have been recently developed in each family: one family has been described by Chuenkongkaew et al. (2005) (F19) and the other was F09 (Fig. 1a) presented in this study. In each of these two families, the G11778A mutation load was 18% in the blood of the maternal ancestor, but it was not detectable at all in the blood of her siblings, indicating that the mutation may be a relatively new occurrence in the germ-line of their mother. However, another possibility would be that the mutation had existed before but did not segregate to her siblings (Howell et al. 2005). MtDNA samples from other informative maternal relatives would be required to confirm the two de novo occurrences of the mutation.
An observation of the three large heteroplasmic pedigrees revealed that the mutation load of the heteroplasmic G11778A mutation in the leukocytes tented to increase in subsequent generations (Fig. 1a–c), thereby supporting the hypothesis of a selection process in favour of the mutated allele. We then analysed the mother-offspring heteroplasmic transmission in other eight heteroplasmic families from our collection. All of the affected probands in these families were omitted from the analysis to reduce ascertainment bias, as suggested by Chinnery et al. (2001), leaving 37 mother-offspring transmissions in which we compared the degree of blood heteroplasmy in the mothers and offspring. An offspring was classified as harbouring a higher or lower mutation load than his/her mother if his/her mutation load was 5% higher or 5% lower than that of his/her mother, respectively. This analysis revealed that 70% (26/37) of the offspring had a higher mutation load than their mothers, while only 16% (6/37) had a relatively lower mutation load; the mutation load of the remaining 14% (5/37) was approximately the same as that of their mothers. The average mutation load in the mothers (n=17) was 44.6±26.1% (median: 44%), while that in the offspring (n = 37) was 64.4±30.1% (median: 70%). These results are in accordance with those of several other studies (Bolhuis et al. 1990; Howell et al. 1994; Jacobi et al. 2001; Savontaus 1995; Smith et al. 1993), but not with a recent analysis of data from published pedigrees by Chinnery et al. (2001).
Given that the degree of heteroplasmy influences the expression of this disease, one argument often used against the hypothesized mechanism of selective transmission is an ascertainment bias: i.e. the heteroplasmic pedigrees were ascertained through the affected probands who should have a high mutation load. We therefore reduced the ascertainment bias by excluding the probands from the analysis. In addition, the removal of all affected offspring from the analysis still resulted in only 19% (6/31) of the offspring having a lower mutation load than their mothers. Therefore, our results should not be severely biased by the sampling method through affected patients.
We performed further analysis with three informative heteroplasmic pedigrees (F09, F19 and F24). Each pedigree was analysed separately, starting with one heteroplasmic mother in generation 2 (Fig. 2). When the average degree of heteroplasmy was compared with those of the next generations (generations 3 and 4), it was found to be significantly higher (P=0.005; ANOVA) in F19, the heteroplasmic family previously reported in Chuenkongkaew et al. (2005). The mean percentages of heteroplasmy of each generation between three generations (generations 2, 3 and 4) of F09 (P=0.103) and F24 (P=0.245) had a pattern similar to that of F19 (Fig. 2), although the difference was not statistically significant. Our data therefore supports the hypothesis that segregation of the G11778A mutation during oogenesis is subject to positive selection rather than being a random process.
Graph showing the average percentage of heteroplasmy of each generation for three pedigrees (F09, F19 and F24), with the starting point being one heteroplasmic mother of each pedigree in generation 2. The average percentage of heteroplasmy of each generation in the same pedigree was compared and found to be significantly increased in F19 (P=0.005) using ANOVA. The average percentage of heteroplasmy of each generation from F09 and F24 showed a similar pattern, although it was not statistically significant (P=0.103 in F09 and P=0.245 in F24)
There were offspring, however, who carried a lower blood mutation load than their mothers, indicating that if selective transmission does actually occur, it is either not total or may be subject to other modifying factors. A recent study shows that there is a directional selection for different mtDNA genotypes in a heteroplasmic mouse model and that the selection is partly controlled by nuclear quantitative loci (Battersby et al. 2003).
Various studies report the rapid segregation of LHON mutations from a low level of heteroplasmy to a practically homoplasmic level in only a few generations (Bolhuis et al. 1990; Howell et al. 1994; Huoponen 2001; Jacobi et al. 2001; Vilkki et al. 1990). This was also observed in our pedigrees, the most evident of which was the shift from 18% (F09, II-1) to homoplasmy (F09, III-10 and III-15) over only one generation (Fig. 1a).
Pedigree F28 (Fig. 1c) was quite interesting in the way that it could clearly be separated into two distinct, homoplasmic (descended from individual II-11) and heteroplasmic (descended from individual II-8) branches. With the exception of showing heteroplasmy for G11778A, two individuals, one from each branch, carried the same mitochondrial genome (Table 1). Analysis of this pedigree clearly revealed that disease penetrance was higher in the homoplasmic branch than in its heteroplasmic counterpart. When only individuals in the maternal line older than 30 years were considered, the estimated penetrance was 50% (5/10) in the homoplasmic branch and 13% (3/23) in the heteroplasmic branch. The other two heteroplasmic families, F09 (Fig. 1a) and F24 (Fig. 1b), also demonstrated a low penetrance of the disease. Only individuals with a high mutation load (more than 85%) in these heteroplasmic families became affected, although some individuals with almost a homoplasmic mutation load in the same pedigree were unaffected.
From the patients in our pedigree collections, the difference in the average final visual acuities (latest logMAR) between heteroplasmic and homoplasmic patients was compared using Mann-Whitney test and found to be significantly different (P=0.023). A Kaplan-Meier survival estimation was carried out to predict the probability of Leber-free survival from two heteroplasmic groups (one group carrying a mutation load between 75 and 95% and the other group carrying mutation load less than 75%) and one homoplasmic group (carrying a mutation load greater than 95%) (Fig. 3). The difference between these three groups was significantly different (P<0.0005; log-rank test for equality of survivor functions). These results on the pedigrees and analyses clearly supported the hypothesis that heteroplasmy is one of the factors influencing disease penetrance.
A Kaplan-Meier survival estimate using software Intercooled Stata 9.1 for Windows was performed between the probability of Leber-free survival and age at onset for three groups of patients: group 1, carrying a greater than 95% mutation load; group 2, carrying a mutation load between 75 and 95%; group 3, carrying a less than 75% mutation load. The difference between these three groups was compared by a log-rank test for equality of survivor functions and found to be significant (P<0.0005)
Low-penetrance branches
Within the framework of the biological phenomenon of low-penetrance branching in LHON pedigrees, as proposed by Howell and Mackey (1998), homoplasmic pedigree F30 (Fig. 1d) was of great interest as it was quite clear that all of the affected members of the family clustered in only one branch that descended from individual II-2. In comparison, those branches descended from individuals II-5 and II-8 contained no affected family members among 35 maternal relatives spanning three generations, despite the fact that all ten individuals tested carried the G11778A mutation in a homoplasmic fashion. The results of the analysis of this large, extended LHON family supported the premise that “low-penetrance branching in LHON matrilineal pedigrees is a biologically real phenomenon” (Howell and Mackey 1998) that cannot be explained by the G11778A mutation itself. Therefore, there must be other factors interacting with the G11778A mutation and influencing disease penetrance. Since the low-penetrance branches possessed the same mtDNA nucleotide variants as the relatively high-penetrance branch, as confirmed by total mitochondrial sequence (Table 1), one possibility is that the paternal nuclear allele(s) might be involved in promoting or suppressing disease expression in the offspring. Alternatively, environmental factors, such as chemical exposure, alcohol drinking or smoking, may contribute to the difference in disease penetrance.
References
Battersby BJ, Loredo-Osti JC, Shoubridge EA (2003) Nuclear genetic control of mitochondrial DNA segregation. Nat Genet 33:183–186
Bolhuis PA, Bleeker-Wagemakers EM, Ponne NJ, Van Schooneveld MJ, Westerveld A, Van den Bogert C, Tabak HF (1990) Rapid shift in genotype of human mitochondrial DNA in a family with Leber’s hereditary optic neuropathy. Biochem Biophys Res Commun 170:994–997
Chinnery PF, Andrews RM, Turnbull DM, Howell NN (2001) Leber hereditary optic neuropathy: does heteroplasmy influence the inheritance and expression of the G11778A mitochondrial DNA mutation? Am J Med Genet 98:235–243
Chuenkongkaew WL, Lertrit P, Poonyathalang A, Sura T, Ruangvaravate N, Atchaneeyasakul L, Suphavilai R (2001) Leber’s hereditary optic neuropathy in Thailand. Jpn J Ophthalmol 45:665–668
Chuenkongkaew WL, Lertrit P, Limwongse C, Nilanont Y, Boonyapisit K, Sangruchi T, Chirapapaisan N, Suphavilai R (2005) An unusual family with Leber’s hereditary optic neuropathy and facioscapulohumeral muscular dystrophy. Eur J Neurol 12:388–391
Ghosh SS, Fahy E, Bodis-Wollner I, Sherman J, Howell N (1996) Longitudinal study of a heteroplasmic 3460 Leber hereditary optic neuropathy family by multiplexed primer-extension analysis and nucleotide sequencing. Am J Hum Genet 58:325–334
Harding AE, Sweeney MG, Govan GG, Riordan-Eva P (1995) Pedigree analysis in Leber hereditary optic neuropathy families with a pathogenic mtDNA mutation. Am J Hum Genet 57:77–86
Howell N, Mackey DA (1998) Low-penetrance branches in matrilineal pedigrees with Leber hereditary optic neuropathy. Am J Hum Genet 63:1220–1224
Howell N, Xu M, Halvorson S, Bodis-Wollner I, Sherman J (1994) A heteroplasmic LHON family: tissue distribution and transmission of the 11778 mutation. Am J Hum Genet 55:203–206
Howell N, Kubacka I, Keers SM, Turnbull DM, Chinnery PF (2005) Co-segregation and heteroplasmy of two coding-region mtDNA mutations within a matrilineal pedigree. Hum Genet 116:28–32
Huoponen K (2001) Leber hereditary optic neuropathy: clinical and molecular genetic findings. Neurogenetics 3:119–125
Jacobi FK, Leo-Kottler B, Mittelviefhaus K, Zrenner E, Meyer J, Pusch CM, Wissinger B (2001) Segregation patterns and heteroplasmy prevalence in Leber’s hereditary optic neuropathy. Invest Ophthalmol Vis Sci 42:1208–1214
Lertrit P, Ruangvaravate N, Trongpanich Y, Imsumran A, Mungkornkarn C, Neungton N (1999) Leber’s hereditary optic neuropathy (LHON) with mitochondrial ND4 gene mutation (11778) in a Thai patient. J Med Assoc Thai 82:59–64
Mackey DA, Oostra RJ, Rosenberg T, Nikoskelainen E, Bronte-Stewart J, Poulton J, Harding AE, Govan G, Bolhuis PA, Norby S (1996) Primary pathogenic mtDNA mutations in multigeneration pedigrees with Leber hereditary optic neuropathy. Am J Hum Genet 59:481–485
Man PY, Turnbull DM, Chinnery PF (2002) Leber hereditary optic neuropathy. J Med Genet 39:162–169
Man PY, Griffiths PG, Brown DT, Howell N, Turnbull DM, Chinnery PF (2003) The epidemiology of Leber hereditary optic neuropathy in the North East of England. Am J Hum Genet 72:333–339
Newman NJ, Lott MT, Wallace DC (1991) The clinical characteristics of pedigrees of Leber’s hereditary optic neuropathy with the 11778 mutation. Am J Ophthalmol 111:750–762
Nikoskelainen EK, Savontaus ML, Wanne OP, Katila MJ, Nummelin KU (1987) Leber’s hereditary optic neuroretinopathy, a maternally inherited disease. A genealogic study in four pedigrees. Arch Ophthalmol 105:665–671
Phasukkijwatana N, Chuenkongkaew WL, Suphavilai R, Suktitipat B, Pingsuthiwong S, Ruangvaravate N, Atchaneeyasakul LO, Warrasak S, Poonyathalang A, Sura T, Lertrit P (2006) The unique characteristics of Thai Leber hereditary optic neuropathy: analysis of 30 G11778A pedigrees. J Hum Genet 81:298–304
Riordan-Eva P, Sanders MD, Govan GG, Sweeney MG, Da Costa J, Harding AE (1995) The clinical features of Leber’s hereditary optic neuropathy defined by the presence of a pathogenic mitochondrial DNA mutation. Brain 118:319–337
Savontaus ML (1995) mtDNA mutations in Leber’s hereditary optic neuropathy. Biochim Biophys Acta 1271:261–263
Smith KH, Johns DR, Heher KL, Miller NR (1993) Heteroplasmy in Leber’s hereditary optic neuropathy. Arch Ophthalmol 111:1486–1490
Tharaphan P, Chuenkongkaew WL, Luangtrakool K, Sanpachudayan T, Suktitipat B, Suphavilai R, Srisawat C, Sura T, Lertrit P (2006) Mitochondrial DNA haplogroup distribution in pedigrees of Southeast Asian G11778A Leber hereditary optic neuropathy. J Neuro-Ophthalmol (in press)
Vilkki J, Savontaus ML, Nikoskelainen EK (1990) Segregation of mitochondrial genomes in a heteroplasmic lineage with Leber hereditary optic neuroretinopathy. Am J Hum Genet 47:95–100
Acknowledgements
The authors would like to thank Pattamon Tharaphan, Thitima Sanpachudayan and Sarinee Pingsutthiwong for their great assistance in the field trip, Benjamas Intharabut and Treenud Suntisiri for their help in the DNA extraction, Yutthana Joijinda for sequencing the entire mitochondrial genome and Supannee Kaewsutthi and Preeyaphan Meesap for their help in preparing the manuscript. We would also like to thank Dr. Chayanon Peerapittayamongkol for his valuable suggestions with respect to the statistical analysis. This work was supported by the Thailand Research Fund (TRF) grant no. BRG4580018 to Lertrit P.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Phasukkijwatana, N., Chuenkongkaew, W.L., Suphavilai, R. et al. Transmission of heteroplasmic G11778A in extensive pedigrees of Thai Leber hereditary optic neuropathy. J Hum Genet 51, 1110–1117 (2006). https://doi.org/10.1007/s10038-006-0073-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10038-006-0073-6
Keywords
This article is cited by
-
Late-onset Leber hereditary optic neuropathy mimicking Susac’s syndrome
Journal of Neurology (2010)