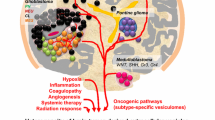
Abstract
To promote the tumor growth, angiogenesis, metabolism, and invasion, glioblastoma (GBM) cells subvert the surrounding microenvironment by influencing the endogenous activity of other brain cells including endothelial cells, macrophages, astrocytes, and microglia. Large number of studies indicates that the intra-cellular communication between the different cell types of the GBM microenvironment occurs through the functional transfer of oncogenic components such as proteins, non-coding RNAs, DNA and lipids via the release and uptake of extracellular vesicles (EVs). Unlike the communication through the secretion of chemokines and cytokines, the transfer and gene silencing activity of microRNAs through EVs is more complex as the biogenesis and proper packaging of microRNAs is crucial for their uptake by recipient cells. Although the specific mechanism of EV-derived microRNA uptake and processing in recipient cells is largely unknown, the screening, identifying and finally targeting of the EV-associated pro-tumorigenic microRNAs are emerging as new therapeutic strategy to combat the GBM.

Similar content being viewed by others
References
Stupp R et al (2009) Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol 10(5):459–466
Ostrom QT et al (2014) CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2007–2011. Neuro Oncol 16(Suppl 4):iv1–iv63
Huntzinger E, Izaurralde E (2011) Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet 12(2):99–110
Piwecka M et al (2015) Comprehensive analysis of microRNA expression profile in malignant glioma tissues. Mol Oncol 9(7):1324–1340
Lee Y et al (2004) MicroRNA genes are transcribed by RNA polymerase II. EMBO J 23(20):4051–4060
Denli AM et al (2004) Processing of primary microRNAs by the microprocessor complex. Nature 432(7014):231–235
Gregory RI et al (2004) The microprocessor complex mediates the genesis of microRNAs. Nature 432(7014):235–240
Bohnsack MT, Czaplinski K, Gorlich D (2004) Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA 10(2):185–191
Chendrimada TP et al (2005) TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436(7051):740–744
Chan JA, Krichevsky AM, Kosik KS (2005) MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res 65(14):6029–6033
Ciafre SA et al (2005) Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun 334(4):1351–1358
Furnari FB et al (2007) Malignant astrocytic glioma: genetics, biology, and paths to treatment. Genes Dev 21(21):2683–2710
Sumazin P et al (2011) An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell 147(2):370–381
Brennan CW et al (2013) The somatic genomic landscape of glioblastoma. Cell 155(2):462–477
Phillips HS et al (2006) Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9(3):157–173
Verhaak RG et al (2010) Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17(1):98–110
Kim TM et al (2011) A developmental taxonomy of glioblastoma defined and maintained by MicroRNAs. Cancer Res 71(9):3387–3399
Tang W et al (2013) Subtyping glioblastoma by combining miRNA and mRNA expression data using compressed sensing-based approach. EURASIP J Bioinf Syst Biol 2013(1):2
Genovese G et al (2012) microRNA regulatory network inference identifies miR-34a as a novel regulator of TGF-beta signaling in glioblastoma. Cancer Discov 2(8):736–749
Mathew LK et al (2014) miR-218 opposes a critical RTK-HIF pathway in mesenchymal glioblastoma. Proc Natl Acad Sci USA 111(1):291–296
Wang H et al (2015) Upregulation of miR-181s reverses mesenchymal transition by targeting KPNA4 in glioblastoma. Sci Rep 5:13072
Hayes J et al (2015) Prediction of clinical outcome in glioblastoma using a biologically relevant nine-microRNA signature. Mol Oncol 9(3):704–714
Papagiannakopoulos T et al (2012) Pro-neural miR-128 is a glioma tumor suppressor that targets mitogenic kinases. Oncogene 31(15):1884–1895
Cancer Genome Atlas Research N (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455(7216):1061–1068
Wang F et al (2014) MiRNA-181c inhibits EGFR-signaling-dependent MMP9 activation via suppressing Akt phosphorylation in glioblastoma. Tumour Biol 35(9):8653–8658
Kefas B et al (2008) microRNA-7 inhibits the epidermal growth factor receptor and the Akt pathway and is down-regulated in glioblastoma. Cancer Res 68(10):3566–3572
Silber J et al (2008) miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med 6(1):1–17
Yin D et al (2013) miR-34a functions as a tumor suppressor modulating EGFR in glioblastoma multiforme. Oncogene 32(9):1155–1163
Kim J et al (2014) microRNA-148a is a prognostic oncomiR that targets MIG6 and BIM to regulate EGFR and apoptosis in glioblastoma. Cancer Res 74(5):1541–1553
Lu J et al (2005) MicroRNA expression profiles classify human cancers. Nature 435(7043):834–838
Chen Y et al (2008) MicroRNA-21 down-regulates the expression of tumor suppressor PDCD4 in human glioblastoma cell T98G. Cancer Lett 272(2):197–205
Gabriely G et al (2008) MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol 28(17):5369–5380
Corsten MF et al (2007) MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell-delivered S-TRAIL in human gliomas. Cancer Res 67(19):8994–9000
Kwak HJ et al (2011) Downregulation of Spry2 by miR-21 triggers malignancy in human gliomas. Oncogene 30(21):2433–2442
Sasayama T et al (2009) MicroRNA-10b is overexpressed in malignant glioma and associated with tumor invasive factors, uPAR and RhoC. Int J Cancer 125(6):1407–1413
Gabriely G et al (2011) Human glioma growth is controlled by microRNA-10b. Cancer Res 71(10):3563–3572
Lavon I et al (2010) Gliomas display a microRNA expression profile reminiscent of neural precursor cells. Neuro Oncol 12(5):422–433
Ernst A et al (2010) De-repression of CTGF via the miR-17-92 cluster upon differentiation of human glioblastoma spheroid cultures. Oncogene 29(23):3411–3422
Dews M et al (2010) The myc-miR-17~92 axis blunts TGF{beta} signaling and production of multiple TGF{beta}-dependent antiangiogenic factors. Cancer Res 70(20):8233–8246
Kim H et al (2010) Integrative genome analysis reveals an oncomir/oncogene cluster regulating glioblastoma survivorship. Proc Natl Acad Sci 107(5):2183–2188
Galli R et al (2004) Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res 64(19):7011–7021
Singh SK et al (2004) Identification of human brain tumour initiating cells. Nature 432(7015):396–401
Yuan X et al (2004) Isolation of cancer stem cells from adult glioblastoma multiforme. Oncogene 23(58):9392–9400
Bao S et al (2006) Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444(7120):756–760
Bier A et al (2013) MicroRNA-137 is downregulated in glioblastoma and inhibits the stemness of glioma stem cells by targeting RTVP-1. Oncotarget 4(5):665–676
Sathyan P et al (2015) Mir-21–Sox2 axis delineates glioblastoma subtypes with prognostic impact. J Neurosci 35(45):15097–15112
Lopez-Bertoni H et al (2015) DNMT-dependent suppression of microRNA regulates the induction of GBM tumor-propagating phenotype by Oct4 and Sox2. Oncogene 34(30):3994–4004
Li Y et al (2009) MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res 69(19):7569–7576
Kefas B et al (2009) The neuronal microRNA miR-326 acts in a feedback loop with notch and has therapeutic potential against brain tumors. J Neurosci 29(48):15161–15168
Floyd DH et al (2014) Novel anti-apoptotic microRNAs 582-5p and 363 promote human glioblastoma stem cell survival via direct inhibition of caspase 3, caspase 9, and Bim. PLoS One 9(5):e96239
Godlewski J et al (2008) Targeting of the Bmi-1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res 68(22):9125–9130
Peruzzi P et al (2013) MicroRNA-128 coordinately targets polycomb repressor complexes in glioma stem cells. Neuro Oncol 15(9):1212–1224
Wong HK et al (2015) The cancer genome atlas analysis predicts MicroRNA for targeting cancer growth and vascularization in glioblastoma. Mol Ther 23(7):1234–1247
Alvarado AG et al (2015) Coordination of self-renewal in glioblastoma by integration of adhesion and microRNA signaling. Neuro Oncol. doi:10.1093/neuonc/nov196
Zhang Y et al (2014) Multiple receptor tyrosine kinases converge on microRNA-134 to control KRAS, STAT5B, and glioblastoma. Cell Death Differ 21(5):720–734
Gilbertson RJ, Rich JN (2007) Making a tumour’ bed: glioblastoma stem cells and the vascular niche. Nat Rev Cancer 7(10):733–736
Babae N et al (2014) Systemic miRNA-7 delivery inhibits tumor angiogenesis and growth in murine xenograft glioblastoma. Oncotarget 5(16):6687–6700
Sun J et al (2015) MiR-137 inhibits proliferation and angiogenesis of human glioblastoma cells by targeting EZH2. J Neurooncol 122(3):481–489
Ostergaard L et al (2013) The relationship between tumor blood flow, angiogenesis, tumor hypoxia, and aerobic glycolysis. Cancer Res 73(18):5618–5624
Fang L et al (2011) MicroRNA miR-93 promotes tumor growth and angiogenesis by targeting integrin-beta8. Oncogene 30(7):806–821
Smits M et al (2012) Myc-associated zinc finger protein (MAZ) is regulated by miR-125b and mediates VEGF-induced angiogenesis in glioblastoma. FASEB J 26(6):2639–2647
Bronisz A et al (2014) Extracellular vesicles modulate the glioblastoma microenvironment via a tumor suppression signaling network directed by miR-1. Cancer Res 74(3):738–750
Warburg O (1956) On the origin of cancer cells. Science 123(3191):309–314
Godlewski J et al (2010) MicroRNA-451 regulates LKB1/AMPK signaling and allows adaptation to metabolic stress in glioma cells. Mol Cell 37(5):620–632
Godlewski J et al (2010) microRNA-451: a conditional switch controlling glioma cell proliferation and migration. Cell Cycle 9(14):2742–2748
Ansari KI et al (2015) Glucose-based regulation of miR-451/AMPK signaling depends on the OCT1 transcription factor. Cell Rep 11(6):902–909
Bronisz A, Chiocca EA, Godlewski J (2015) Response to energy depletion: miR-451/AMPK loop. Oncotarget 6(20):17851–17852
Hardie DG (2015) AMPK: positive and negative regulation, and its role in whole-body energy homeostasis. Curr Opin Cell Biol 33:1–7
Kefas B et al (2010) Pyruvate kinase M2 is a target of the tumor-suppressive microRNA-326 and regulates the survival of glioma cells. Neuro Oncol 12(11):1102–1112
Masui K et al (2013) mTOR complex 2 controls glycolytic metabolism in glioblastoma through FoxO acetylation and upregulation of c-Myc. Cell Metab 18(5):726–739
Charles NA et al (2011) The brain tumor microenvironment. Glia 59(8):1169–1180
Nduom EK, Weller M, Heimberger AB (2015) Immunosuppressive mechanisms in glioblastoma. Neuro Oncol 17(Suppl 7):vii9–vii14
Codo P et al (2014) MicroRNA-mediated down-regulation of NKG2D ligands contributes to glioma immune escape. Oncotarget 5(17):7651–7662
Wei J et al (2013) miR-124 inhibits STAT3 signaling to enhance T cell-mediated immune clearance of glioma. Cancer Res 73(13):3913–3926
Ueda R et al (2009) Dicer-regulated microRNAs 222 and 339 promote resistance of cancer cells to cytotoxic T-lymphocytes by down-regulation of ICAM-1. Proc Natl Acad Sci 106(26):10746–10751
Hussain SF et al (2006) The role of human glioma-infiltrating microglia/macrophages in mediating antitumor immune responses. Neuro Oncol 8(3):261–279
Xu S et al (2014) Effect of miR-142-3p on the M2 macrophage and therapeutic efficacy against murine glioblastoma. J Natl Cancer Inst 106(8):dju162. doi:10.1093/jnci/dju162
Wei J et al (2015) MiR-138 exerts anti-glioma efficacy by targeting immune checkpoints. Neuro Oncol. doi:10.1093/neuonc/nov292
Ohno M et al (2013) Expression of miR-17-92 enhances anti-tumor activity of T-cells transduced with the anti-EGFRvIII chimeric antigen receptor in mice bearing human GBM xenografts. J Immunother Cancer 1:21
Kosaka A et al (2015) Transgene-derived overexpression of miR-17-92 in CD8 + T-cells confers enhanced cytotoxic activity. Biochem Biophys Res Commun 458(3):549–554
Heijnen HF et al (1999) Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood 94(11):3791–3799
Kowal J, Tkach M, Thery C (2014) Biogenesis and secretion of exosomes. Curr Opin Cell Biol 29:116–125
Dainiak N (1991) Surface membrane-associated regulation of cell assembly, differentiation, and growth. Blood 78(2):264–276
Mathivanan S et al (2010) Proteomics analysis of A33 immunoaffinity-purified exosomes released from the human colon tumor cell line LIM1215 reveals a tissue-specific protein signature. Mol Cell Proteom 9(2):197–208
Ostrowski M et al. (2010) Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol 12(1):19–30; sup pp 1–13
Dolo V et al (1998) Selective localization of matrix metalloproteinase 9, beta1 integrins, and human lymphocyte antigen class I molecules on membrane vesicles shed by 8701-BC breast carcinoma cells. Cancer Res 58(19):4468–4474
Schiera G et al (2007) Neurons produce FGF2 and VEGF and secrete them at least in part by shedding extracellular vesicles. J Cell Mol Med 11(6):1384–1394
Proia P et al (2008) Astrocytes shed extracellular vesicles that contain fibroblast growth factor-2 and vascular endothelial growth factor. Int J Mol Med 21(1):63–67
Li CC et al (2013) Glioma microvesicles carry selectively packaged coding and non-coding RNAs which alter gene expression in recipient cells. RNA Biol 10(8):1333–1344
Hessvik NP et al (2012) Profiling of microRNAs in exosomes released from PC-3 prostate cancer cells. Biochim Biophys Acta 1819(11–12):1154–1163
Armstrong DA et al (2015) MicroRNA molecular profiling from matched tumor and bio-fluids in bladder cancer. Mol Cancer 14(1):194
Villarroya-Beltri C et al (2013) Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun 4:2980
Koppers-Lalic D et al (2014) Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep 8(6):1649–1658
van der Vos KE et al (2016) Directly visualized glioblastoma-derived extracellular vesicles transfer RNA to microglia/macrophages in the brain. Neuro Oncol 18(1):58–69
Skog J et al (2008) Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol 10(12):1470–1476
Ridder K et al (2015) Extracellular vesicle-mediated transfer of functional RNA in the tumor microenvironment. Oncoimmunology 4(6):e1008371
Gasser O et al (2003) Characterisation and properties of ectosomes released by human polymorphonuclear neutrophils. Exp Cell Res 285(2):243–257
Eken C et al (2008) Polymorphonuclear neutrophil-derived ectosomes interfere with the maturation of monocyte-derived dendritic cells. J Immunol 180(2):817–824
Pluskota E et al (2008) Expression, activation, and function of integrin alphaMbeta2 (Mac-1) on neutrophil-derived microparticles. Blood 112(6):2327–2335
Hoshino A et al (2015) Tumour exosome integrins determine organotropic metastasis. Nature 527(7578):329–335
Gasser O, Schifferli JA (2004) Activated polymorphonuclear neutrophils disseminate anti-inflammatory microparticles by ectocytosis. Blood 104(8):2543–2548
Al-Nedawi K et al (2008) Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol 10(5):619–624
Nakase I et al (2015) Active macropinocytosis induction by stimulation of epidermal growth factor receptor and oncogenic Ras expression potentiates cellular uptake efficacy of exosomes. Sci Rep 5:10300
Svensson KJ et al (2013) Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid Raft-mediated endocytosis negatively regulated by caveolin-1. J Biol Chem 288(24):17713–17724
Christianson HC et al (2013) Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc Natl Acad Sci USA 110(43):17380–17385
Atai NA et al (2013) Heparin blocks transfer of extracellular vesicles between donor and recipient cells. J Neurooncol 115(3):343–351
Montecalvo A et al (2012) Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 119(3):756–766
Parolini I et al (2009) Microenvironmental pH is a key factor for exosome traffic in tumor cells. J Biol Chem 284(49):34211–34222
Mineo M et al (2012) Exosomes released by K562 chronic myeloid leukemia cells promote angiogenesis in a Src-dependent fashion. Angiogenesis 15(1):33–45
Kucharzewska P et al (2013) Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc Natl Acad Sci USA 110(18):7312–7317
Agrawal R et al (2014) Hypoxic signature of microRNAs in glioblastoma: insights from small RNA deep sequencing. BMC Genom 15:686
Tadokoro H et al (2013) Exosomes derived from hypoxic leukemia cells enhance tube formation in endothelial cells. J Biol Chem 288(48):34343–34351
Li R et al (2014) Identification of intrinsic subtype-specific prognostic microRNAs in primary glioblastoma. J Exp Clin Cancer Res 33:9
Yan W et al (2014) MicroRNA expression patterns in the malignant progression of gliomas and a 5-microRNA signature for prognosis. Oncotarget 5(24):12908–12915
Hu J et al (2016) MiR-215 is induced post-transcriptionally via HIF-Drosha complex and mediates glioma-initiating cell adaptation to hypoxia by targeting KDM1B. Cancer Cell 29(1):49–60
Chen CC et al (2009) Stereotactic brain biopsy: single center retrospective analysis of complications. Clin Neurol Neurosurg 111(10):835–839
Westphal M, Lamszus K (2015) Circulating biomarkers for gliomas. Nat Rev Neurol 11(10):556–566
Pisitkun T, Shen RF, Knepper MA (2004) Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci USA 101(36):13368–13373
Gallo A et al (2012) The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One 7(3):e30679
Akers JC et al (2013) MiR-21 in the extracellular vesicles (EVs) of cerebrospinal fluid (CSF): a platform for glioblastoma biomarker development. PLoS One 8(10):e78115
Qu S, Guan J, Liu Y (2015) Identification of microRNAs as novel biomarkers for glioma detection: a meta-analysis based on 11 articles. J Neurol Sci 348(1–2):181–187
Shi L et al (2010) MiR-21 protected human glioblastoma U87MG cells from chemotherapeutic drug temozolomide induced apoptosis by decreasing Bax/Bcl-2 ratio and caspase-3 activity. Brain Res 1352:255–264
Teplyuk NM et al (2012) MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro Oncol 14(6):689–700
Roth P et al (2011) A specific miRNA signature in the peripheral blood of glioblastoma patients. J Neurochem 118(3):449–457
Manterola L et al (2014) A small noncoding RNA signature found in exosomes of GBM patient serum as a diagnostic tool. Neuro Oncol 16(4):520–527
Bronisz A et al (2012) Reprogramming of the tumour microenvironment by stromal PTEN-regulated miR-320. Nat Cell Biol 14(2):159–167
Sun JY et al (2015) MicroRNA-320 inhibits cell proliferation in glioma by targeting E2F1. Mol Med Rep 12(2):2355–2359
Financial support
NCI 1R01 CA176203-01A1 (JG).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declare no conflict of interest.
Additional information
A. K. Rooj and M. Mineo contributed equally to the manuscript.
Rights and permissions
About this article
Cite this article
Rooj, A.K., Mineo, M. & Godlewski, J. MicroRNA and extracellular vesicles in glioblastoma: small but powerful. Brain Tumor Pathol 33, 77–88 (2016). https://doi.org/10.1007/s10014-016-0259-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10014-016-0259-3