Abstract
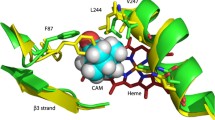
Cytochrome P450 enzymes are hemoprotein monooxygenases that catalyse the oxidation of a variety of compounds. The mechanism by which camphor, the natural substrate of Cytochrome P450cam (P450cam), accesses the active site is a long-standing puzzle, although putative access channels have been proposed. A thermal motion pathway (TMP) analysis was performed on the crystal structure of P450cam with camphor bound. Hereby, three distinct thermal motion pathway families (TMPFs) were found. Possible substrate access channels obtained by this analysis based on B-factors are compared with exit channels explored by molecular dynamics simulations (MDS) by imposing an artificial expulsion force on the substrate in addition to the standard MD force field. Two out of three TMPFs are supported by results obtained with the random expulsion MDS method. However, the pathway found by the TMP method to have the highest average B-factor could not be observed by MDS. The pathway proposed from crystallographic data, which is a small opening above the active site located near residues 185, 87 and 395 corresponds to the TMPF with the second highest average B-factor.
Similar content being viewed by others
Author information
Authors and Affiliations
Additional information
Received: 12 June 1997 / Accepted: 3 July 1997 / Published: 15 August 1997
Rights and permissions
About this article
Cite this article
Lüdemann, S., Carugo, O. & Wade, R. Substrate Access to Cytochrome P450cam: A Comparison of a Thermal Motion Pathway Analysis with Molecular Dynamics Simulation Data. J Mol Med 3, 369–374 (1997). https://doi.org/10.1007/s008940050053
Issue Date:
DOI: https://doi.org/10.1007/s008940050053