Abstract
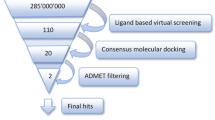
Alzheimer’s disease (AD) is a chronic neurodegenerative disease of the elderly that seriously affects the quality of life and the life expectancy of those affected. There is, as yet, no effective drug treatment of AD, although several acetylcholinesterase (AChE) inhibitors and a glutamate antagonist can provide relief from its symptoms. Recent studies have indicated that the overactivation of poly(ADP-ribose) polymerase-1 (PARP-1) may promote nerve cell death in the brains of AD patients, implying that PARP-1 inhibition may have therapeutic value for the treatment of AD. Therefore, it is important to investigate novel agents with both AChE- and PARP-1-inhibitory bioactivities. In this study, the structure-based virtual screening of PARP-1 inhibitors was performed to search for potential agents with high affinities for AChE. The dynamic stability of the selected AChE–ligand complexes was investigated by molecular dynamics (MD) simulation. Two compounds, CID57390505 and CID71605390, showed high affinities for and stability in complex with AChE in docking and MD simulations. Thus, our in silico research identified two compounds with AChE and PARP-1 dual-targeted activities, indicating that this technique could aid attempts to develop more potent agents against AD.






Similar content being viewed by others
References
Prince MJ, Wimo A, Guerchet MM, Ali GC, Wu Y-T, Prina M (2015) Alzheimer report 2015—the global impact of dementia: an analysis of prevalence, incidence, cost and trends. Alzheimer’s Disease International, London
Raskin J, Cummings J, Hardy J, Schuh K, Dean RA (2015) Neurobiology of Alzheimer’s disease: integrated molecular, physiological, anatomical, biomarker, and cognitive dimensions. Curr Alzheimer Res 12(8):712–722
Mangialasche F, Solomon A, Winblad B, Mecocci P, Kivipelto M (2010) Alzheimer’s disease: clinical trials and drug development. Lancet Neurol 9(7):702–716
Zhao FC, Wu Y, Song XJ (2017) Design and development of a novel chalcone derivative as an anticholinesterase inhibitor for possible treatment of dementia. Med Sci Monit 23:3311–3317
Bouchard VJ, Rouleau M, Poirier GG (2003) PARP-1, a determinant of cell survival in response to DNA damage. Exp Hematol 31(6):446–454
Kim MY, Mauro S, Gevry N, Lis JT, Kraus WL (2004) NAD+-dependent modulation of chromatin structure and transcription by nucleosome binding properties of PARP-1. Cell 119(6):803–814
Kim MY, Zhang T, Kraus WL (2005) Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev 19(17):1951–1967
David KK, Andrabi SA, Dawson TM, Dawson VL (2009) Parthanatos, a messenger of death. Front Biosci 14:1116–1128
Wang Y, An R, Umanah GK, Park H, Nambiar K, Eacker SM, Kim B, Bao L, Harraz MM, Chang C, Chen R, Wang JE, Kam TI, Jeong JS, Xie Z, Neifert S, Qian J, Andrabi SA, Blackshaw S, Zhu H, Song H, Ming GL, Dawson VL, Dawson TM (2016) A nuclease that mediates cell death induced by DNA damage and poly(ADP-ribose) polymerase-1. Science 354(6308):aad6872
Andrabi SA, Kim NS, Yu SW, Wang H, Koh DW, Sasaki M, Klaus JA, Otsuka T, Zhang Z, Koehler RC, Hurn PD, Poirier GG, Dawson VL, Dawson TM (2006) Poly(ADP-ribose) (PAR) polymer is a death signal. Proc Natl Acad Sci U S A 103(48):18308–18313
Yu SW, Andrabi SA, Wang H, Kim NS, Poirier GG, Dawson TM, Dawson VL (2006) Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc Natl Acad Sci U S A 103(48):18314–18319
Yu SW, Wang H, Poitras MF, Coombs C, Bowers WJ, Federoff HJ, Poirier GG, Dawson TM, Dawson VL (2002) Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science 297(5579):259–263
Chiarugi A, Moskowitz MA (2003) Poly(ADP-ribose) polymerase-1 activity promotes NF-kappaB-driven transcription and microglial activation: implication for neurodegenerative disorders. J Neurochem 85(2):306–317
Chaitanya GV, Steven AJ, Babu PP (2010) PARP-1 cleavage fragments: signatures of cell-death proteases in neurodegeneration. Cell Commun Signal 8:31
Fatokun AA, Dawson VL, Dawson TM (2014) Parthanatos: mitochondrial-linked mechanisms and therapeutic opportunities. Br J Pharmacol 171(8):2000–2016
Ma Y, Chen H, He X, Nie H, Hong Y, Sheng C, Wang Q, Xia W, Ying W (2012) NAD+ metabolism and NAD(+)-dependent enzymes: promising therapeutic targets for neurological diseases. Curr Drug Targets 13(2):222–229
Ruan Q, Ruan J, Zhang W, Qian F, Yu Z (2018) Targeting NAD+ degradation: the therapeutic potential of flavonoids for Alzheimer's disease and cognitive frailty. Pharmacol Res 128:345–358
Das S, Basu S (2017) Multi-targeting strategies for Alzheimer's disease therapeutics: pros and cons. Curr Top Med Chem 17(27):3017–3061
Lin H, Li Q, Gu K, Zhu J, Jiang X, Chen Y, Sun H (2017) Design of multi-target agents for the treatment of Alzheimer's disease based on tacrine. Curr Top Med Chem 17(27):3000–3016
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (2001) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46(1-3):3–26
Bruns RF, Watson IA (2012) Rules for identifying potentially reactive or promiscuous compounds. J Med Chem 55(22):9763–9772
Dahlin JL, Nissink JW, Strasser JM, Francis S, Higgins L, Zhou H, Zhang Z, Walters MA (2015) PAINS in the assay: chemical mechanisms of assay interference and promiscuous enzymatic inhibition observed during a sulfhydryl-scavenging HTS. J Med Chem 58(5):2091–2113
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) Open Babel: an open chemical toolbox. J Cheminform 3:33
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and testing of a general Amber force field. J Comput Chem 25(9):1157–1174
Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461
Ravindranath PA, Forli S, Goodsell DS, Olson AJ, Sanner MF (2015) AutoDockFR: advances in protein–ligand docking with explicitly specified binding site flexibility. PLoS Comput Biol 11(12):e1004586
Pavelka A, Chovancova E, Damborsky J (2009) HotSpot wizard: a web server for identification of hot spots in protein engineering. Nucleic Acids Res 37(Web Server issue):W376–W383
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30(16):2785–2791
Abraham MJ, Murtola T, Schulz R, Páll S, Smith JC, Hess B, Lindahl E (2015) GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2(C):19–25
Schuttelkopf AW, van Aalten DM (2004) PRODRG: a tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallogr D Biol Crystallogr 60(Pt 8):1355–1363
Kumari R, Kumar R, Open Source Drug Discovery C, Lynn A (2014) g_mmpbsa—a GROMACS tool for high-throughput MM-PBSA calculations. J Chem Inf Model 54(7):1951–1962
Dunn D, Husten J, Ator MA, Chatterjee S (2012) Novel poly(ADP-ribose) polymerase-1 inhibitors. Bioorg Med Chem Lett 22(1):222–224
Desai BS, Monahan AJ, Carvey PM, Hendey B (2007) Blood–brain barrier pathology in Alzheimer’s and Parkinson’s disease: implications for drug therapy. Cell Transplant 16(3):285–299
Ren Y, Houghton PJ, Hider RC, Howes MJ (2004) Novel diterpenoid acetylcholinesterase inhibitors from Salvia miltiorhiza. Planta Med 70(3):201–204
Pinho BR, Ferreres F, Valentao P, Andrade PB (2013) Nature as a source of metabolites with cholinesterase-inhibitory activity: an approach to Alzheimer’s disease treatment. J Pharm Pharmacol 65(12):1681–1700
Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I (1991) Atomic structure of acetylcholinesterase from Torpedo californica: a prototypic acetylcholine-binding protein. Science 253(5022):872–879
Lin G, Lai CY, Liao WC (1999) Molecular recognition by acetylcholinesterase at the peripheral anionic site: structure–activity relationships for inhibitions by aryl carbamates. Bioorg Med Chem 7(12):2683–2689
Hirashima A, Kuwano E, Eto M (2000) Docking study of enantiomeric fonofos oxon bound to the active site of Torpedo californica acetylcholinesterase. Bioorg Med Chem 8(3):653–656
Cheung J, Rudolph MJ, Burshteyn F, Cassidy MS, Gary EN, Love J, Franklin MC, Height JJ (2012) Structures of human acetylcholinesterase in complex with pharmacologically important ligands. J Med Chem 55(22):10282–10286
Ashani Y, Grunwald J, Kronman C, Velan B, Shafferman A (1994) Role of tyrosine 337 in the binding of huperzine A to the active site of human acetylcholinesterase. Mol Pharmacol 45(3):555–560
Van de Waterbeemd H, Camenisch G, Folkers G, Chretien JR, Raevsky OA (1998) Estimation of blood–brain barrier crossing of drugs using molecular size and shape, and H-bonding descriptors. J Drug Target 6(2):151–165
Ghose AK, Herbertz T, Hudkins RL, Dorsey BD, Mallamo JP (2012) Knowledge-based, central nervous system (CNS) lead selection and lead optimization for CNS drug discovery. ACS Chem Neurosci 3:50–68
Ertl P, Rohde B, Selzer P (2000) Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J Med Chem 43:3714–3717
Müller J, Martins A, Csábi J, Fenyvesi F, Könczöl Á, Hunyadi A, Balogh GT (2017) BBB penetration-targeting physicochemical lead selection: ecdysteroids as chemo-sensitizers against CNS tumors. Eur J Pharm Sci 96:571–577
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Availability of data and materials
PARP-1 inhibitors were identified and their physical and chemical properties were obtained from the Pubchem database, https://pubchem.ncbi.nlm.nih.gov, on 7-1-2017. The PDB files were obtained from the RCSB Protein Data Bank on 7-1-2017.
Electronic supplementary material
ESM 1
(XLS 137 kb)
Rights and permissions
About this article
Cite this article
Hu, XM., Dong, W., Cui, ZW. et al. In silico identification of AChE and PARP-1 dual-targeted inhibitors of Alzheimer’s disease. J Mol Model 24, 151 (2018). https://doi.org/10.1007/s00894-018-3696-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-018-3696-6