Abstract
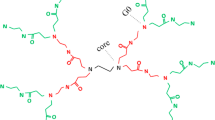
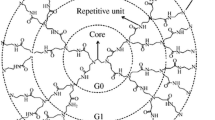
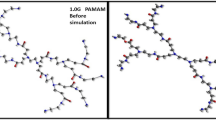
Poly(L-lysine) (PLL) dendrimer are amino acid based macromolecules and can be used as drug delivery agents. Their branched structure allows them to be functionalized by various groups to encapsulate drug agents into their structure. In this work, at first, an attempt was made on all-atom simulation of PLL dendrimer of different generations. Based on all-atom results, a course-grained model of this dendrimer was designed and its parameters were determined, to be used for simulation of three generations of PLL dendrimer, at two pHs. Similar to the all-atom, the coarse-grained results indicated that by increasing the generation, the dendrimer becomes more spherical. At pH 7, the dendrimer had larger size, whereas at pH 12, due to back folding of branching chains, they had the tendency to penetrate into the inner layers. The calculated radial probability and radial distribution functions confirm that at pH 7, the PLL dendrimer has more cavities and as a result it can encapsulate more water molecules into its inner structure. By calculating the moment of inertia and the aspect ratio, the formation of spherical structure for PLL dendrimer was confirmed.








Similar content being viewed by others
References
Baoukina S, Monticelli L, Tieleman DP (2013) Interaction of pristine and functionalizedcarbon nanotubes with lipid membranes. J Phys Chem B 117:12113–12123
Menjoge AR, Kannan RM, Tomalia DA (2010) Dendrimer-based drug and imaging conjugates: design considerations for nanomedical applications. Drug Discov Today 15:171–185
Yonetake K, Masuko T (1999) Poly(propyleneimine) dendrimers peripherally modified with mesogens. Macromolecular 32:6578–6586
Zhang X, Oulad-abdelghani M, Zelkin AN, Wang Y, Haı Y, Benkirane-jessel N, Mainard D, Voegel J, Caruso F (2010) Poly(L-lysine) nanostructured particles for gene delivery and hormone stimulation. Biomaterials 31:1699–1706
Fox ME, Guillaudeu S, Fre JMJ, Jerger K, Macaraeg N, Szoka FC (2009) Synthesis and in Vivo antitumor efficacy of PEGylated Poly(L-lysine) dendrimer-camptothecin conjugates. Mol Pharm 6:1562–1572
Gillies ER, Frechet JM (2005) Dendrimers and dendritic polymers in drug delivery. Drug Discov Today 10:35–43
Stiriba S, Frey H, Haag R (2002) Dendritic polymers in biomedical applications: From potential to clinical use in diagnostics and therapy. Angew Chem Int Ed 41:1329–1334
Niedrhafner P, Šebestik J, Ježek J (2005) Peptide dendrimers. J Peptide Sci 11:757–788
Boas U, Heegaard PM (2004) Dendrimers in drug research. Chem Soc Rev 33:43–63
Perumal OP, Inapagolla R, Kannan S, Kannan RM (2008) The effect of surface functionality on cellular trafficking of dendrimers. Biomaterials 29:3469–347611
Choi JS, Lee EJ, Choi YH, Jeong YJ, Park JS (1999) Poly(ethylene glycol)-block-poly(L-lysine) dendrimer: novel linear polymer/dendrimer block copolymer forming a spherical water-soluble polyionic complex with DNA. Bioconj Chem 10:62–65
Byrne M, Victory D, Hibbitts A, Lanigan M, Heise A, Cryan S (2013) Molecular weight and architectural dependence of well-defined star-shaped poly(lysine) as a gene delivery vector. Biomater Sci 1:1223–1234
Yevlampieva N, Dobrodumov A, Nazarova O, Okatova O (2012) Hydrodynamic behavior of dendrigraft polylysines in water and dimethylformamide. Poly 4:20–31
Okuda T, Kawakami S, Maeie T, Niidome T (2006) Biodistribution characteristics of amino acid dendrimers and their PEGylated derivatives after intravenous administration. J Contrl Rel 114:69–77
Kaminskas LM, Boyd BJ, Karellas P, Krippner GY, Lessene R, Kelly B, Porter CJH (2008) The impact of molecular weight and PEG chain length on the systemic pharmacokinetics of PEGylated poly L-Lysine dendrimers. Mol Pharm 5:449–463
Okuda T, Sugiyama A, Niidome T, Aoyagi H (2004) Characters of dendritic poly(l-lysine) analogues with the terminal lysines replaced with arginines and histidines as gene carriers in vitro. Biomater 25:537–544
Kaminskas LM, Kelly BD, Mcleod VM, Boyd BJ, Krippner GY, Williams ED, Porter CJH (2009) Pharmacokinetics and tumor disposition of PEGylated, methotrexate conjugated Poly-L-lysine dendrimers. Mol Pharm 6:1190–1204
Rossi J, Boiteau L, Collet H, Tsamba BM, Larcher N (2012) Functionalisation of free amino groups of lysine dendrigraft (DGL) polymers. Tetrahedron Lett 53:2976–2979
Liu Y, Bryantsev VS, Diallo MS, Goddard WA III (2009) PAMAM dendrimers undergo pH responsive conformational changes without swelling. J Am Chem Soc 131:2798–2799
Markelov DA, Falkovich SG, Neelov IM, Ilyash MY, Matveev VV, Lähderanta E, Ingman P, Darinskii AA (2015) Molecular dynamics simulation of spin–lattice NMR relaxation in poly- l -lysine dendrimers: manifestation of the semiflexibility effect. Phys Chem Chem Phys 17:3214–3226
Hong K, Liu Y, Porcar L, Liu D, Gao CY, Smith GS, Herwig KW, Cai S, Li X, Wu B, Chen WR, Liu L (2012) Structural response of polyelectrolyte dendrimer towards molecular protonation: the inconsistency revealed by SANS and NMR. J Phys Condens Matter 24:064116
Amjad-Iranagh GK, Modarress H (2014) Molecular simulation study of PAMAM dendrimer composite membranes. J Mol Model 20:2119–2124
Roberts BP, Scanlon MJ, Krippner GY, Chalmers DK (2009) Molecular dynamics of poly(L-lysine) dendrimers with naphthalene disulfonate Caps. Macromol 42:2775–2783
Neelov IM, Markelov DA, Falkovich SG, Okrugin BM, Darinskii AA (2013) Mathematical simulation of lysine dendrimers: temperature dependences. Poly Sci 55:154–161
Lee H, Choi JS, Larson RG (2011) Molecular dynamics studies of the size and internal structure of the PAMAM dendrimer grafted with arginine and histidine. Macromol 44:8681–8686
Kavyani S, Amjad-iranagh S, Modarress H (2014) Aqueous poly(amidoamine) dendrimer G3 and G4 generations with several interior cores at pHs 5 and 7: a molecular dynamics simulation study. J Phys Chem B 118:3257–3266
Lee H, Larson R (2008) Lipid bilayer curvature and pore formation induced by charged linear polymers and dendrimers: the effect of molecular shape. J Phys Chem B 112:12279–12285
Hess B, Kutzner C (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4:435–447
Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC (2005) GROMACS: fast, flexible, and free. J Comput Chem 26:1701–1718
Malde AK, Zuo L, Breeze M, Stroet M, Poger D, Nair PC, Oostenbrink C, Mark AE (2011) An automated force field topology builder (ATB) and repository: version 1.0. J Chem Theory Comput 7:4026–4037
Bussi G, Donadio D, Parrinello M (2007) Canonical sampling through velocity rescaling. J Chem Phys 126:14101–14107
Parrinello M, Rahman A (1981) Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys 52:7182–7190
Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG (1995) A smooth particle mesh Ewald method. J Chem Phys 103:8577–8593
Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH (2007) The MARTINI force field: coarse grained model for biomolecular simulations. J Phys Chem B 111:7812–7824
Monticelli L, Kandasamy SK, Periole X, Larson RG, Tieleman DP, Marrink SJ (2008) The MARTINI coarse-grained force field: extension to proteins. J Chem Theory and Comput 4:819–834
Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3690
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular Ddnamics. J Mol Graph 14:33–38
Falkovich S, Markelov D, Neelov I, Darinskii A (2013) Are structural properties of dendrimers sensitive to the symmetry of branching? Computer simulation of lysine dendrimers. J Chem Phys 139:64903–64913
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rahimi, A., Amjad-Iranagh, S. & Modarress, H. Molecular dynamics simulation of coarse-grained poly(L-lysine) dendrimers. J Mol Model 22, 59 (2016). https://doi.org/10.1007/s00894-016-2925-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-016-2925-0