Abstract
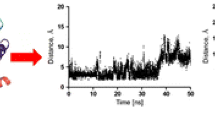
Human dopamine D4 receptor is a GPCR target in the treatment of neurological and psychiatric conditions such as schizophrenia and Parkinson’s disease. The X-ray structure of this receptor has not been resolved so far. Therefore, a proper 3D structure of D4 could provide a good tool in order to design novel ligands against this target. In this study, homology modeling studies were performed to obtain a reasonable structure of the receptor using known templates. The obtained model was subjected to molecular dynamic simulation within a DPPC membrane system. Some structural features of the receptor such as a conserved disulfide bridge and ionic lock were considered in the modeling experiments. The resulted trajectories of simulation were clustered based on the root mean square deviation of the backbone. Some known ligands and decoys were accordingly docked into the representative frames of each cluster. The best final model was finally selected based on its ability to discriminate between active ligands and inactive decoys (ROC = 0.839). The presented model of human D4 receptor could be a promising starting point in future studies of drug design for the described target.

Superposition of human D4 model with the crystal structure of D3 at TM regions










Similar content being viewed by others
References
Fanelli F, De Benedetti PG (2011) Update 1 of: computational modeling approaches to structure–function analysis of G protein-coupled receptors. Chem Rev 111(12):438–535
Bokoch MP, Zou Y, Rasmussen SG, Liu CW, Nygaard R, Rosenbaum DM, Fung JJ, Choi H-J, Thian FS, Kobilka TS (2010) Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor. Nature 463(7277):108–112
Rosano C, Lappano R, Santolla MF, Ponassi M, Donadini A, Maggiolini M (2012) Recent advances in the rationale design of GPER ligands. Curr Med Chem 19(36):6199–6206
Xiang L, Szebeni K, Szebeni A, Klimek V, Stockmeier CA, Karolewicz B, Kalbfleisch J, Ordway GA (2008) Dopamine receptor gene expression in human amygdaloid nuclei: elevated D4 receptor mRNA in major depression. Brain Res 1207:214–224
Beaulieu J-M, Gainetdinov RR (2011) The physiology, signaling, and pharmacology of dopamine receptors. Pharmacol Rev 63(1):182–217
Strange B, Gartmann N, Brenninkmeyer J, Haaker J, Reif A, Kalisch R, Büchel C (2014) Dopamine receptor 4 promoter polymorphism modulates memory and neuronal responses to salience. NeuroImage 84:922–931
Furth KE, Mastwal S, Wang KH, Buonanno A, Vullhorst D (2013) Dopamine, cognitive function, and gamma oscillations: role of D4 receptors. Front Cell Neurosci 7:102. doi:10.3389/fncel.2013.00102
Enguehard-Gueiffier C, Hübner H, El Hakmaoui A, Allouchi H, Gmeiner P, Argiolas A, Melis MR, Gueiffier A (2006) 2-[(4-phenylpiperazin-1-yl) methyl] imidazo (di) azines as selective D4-ligands. Induction of penile erection by 2-[4-(2-methoxyphenyl) piperazin-1-ylmethyl] imidazo [1, 2-a] pyridine (PIP3EA), a potent and selective D4 partial agonist. J Med Chem 49(13):3938–3947
Boyfield I, Brown TH, Coldwell MC, Cooper DG, Hadley MS, Hagan JJ, Healy MA, Johns A, King RJ, Middlemiss DN (1996) Design and synthesis of 2-naphthoate esters as selective dopamine D4 antagonists. J Med Chem 39(10):1946–1948
Berry CB, Bubser M, Jones CK, Hayes JP, Wepy JA, Locuson CW, Daniels JS, Lindsley CW, Hopkins CR (2014) Discovery and characterization of ML398, a potent and selective antagonist of the D4 receptor with in vivo activity. ACS Med Chem Lett 5(9):1060–1064
Sampson D, Zhu XY, Eyunni SVK, Etukala JR, Ofori E, Bricker B, Lamango NS, Setola V, Roth BL, Ablordeppey SY (2014) Identification of a new selective dopamine D4 receptor ligand. Bioorg Med Chem 22(12):3105–3114. doi:10.1016/j.bmc.2014.04.026
Arora J, Bordeleau M, Dube L, Jarvie K, Mazzocco L, Peragine J, Tehim A, Egle I (2005) N-[(3S)-1-Benzylpyrrolidin-3-yl]-(2-thienyl)benzamides: human dopamine D4 ligands with high affinity for the 5-HT2A receptor. Bioorg Med Chem Lett 15(23):5253–5256. doi:10.1016/j.bmcl.2005.08.051
Abdelfattah MAO, Lehmann J, Abadi AH (2013) Discovery of highly potent and selective D4 ligands by interactive SAR study. Bioorg Med Chem Lett 23(18):5077–5081. doi:10.1016/j.bmcl.2013.07.033
Boström J, Böhm M, Gundertofte K, Klebe G (2003) A 3D QSAR study on a set of dopamine D4 receptor antagonists. J Chem Inf Comput Sci 43(3):1020–1027
Fleishman SJ, Unger VM, Ben-Tal N (2006) Transmembrane protein structures without X-rays. Trends Biochem Sci 31(2):106–113. doi:10.1016/j.tibs.2005.12.005
Shahlaei M, Madadkar-Sobhani A, Fassihi A, Saghaie L (2011) Exploring a model of a chemokine receptor/ligand complex in an explicit membrane environment by molecular dynamics simulation: the human CCR1 receptor. J Chem Inf Model 51(10):2717–2730
De Brevern AG (2010) 3D structural models of transmembrane proteins. In: Membrane protein structure determination. Springer, Berlin pp 387–401
Chien EY, Liu W, Zhao Q, Katritch V, Han GW, Hanson MA, Shi L, Newman AH, Javitch JA, Cherezov V (2010) Structure of the human dopamine D3 receptor in complex with a D2/D3 selective antagonist. Science 330(6007):1091–1095
Elbegdorj O, Westkaemper RB, Zhang Y (2013) A homology modeling study toward the understanding of three-dimensional structure and putative pharmacological profile of the G-protein coupled receptor GPR55. J Mol Graph Model 39:50–60
Becker O, Shacham S, Marantz Y, Noiman S (2003) Modeling the 3D structure of GPCRs: advances and application to drug discovery. Current opinion in drug discovery & development 6(3):353–361
Nowak M, Kolaczkowski M, Pawlowski M, Bojarski AJ (2006) Homology modeling of the serotonin 5-HT1A receptor using automated docking of bioactive compounds with defined geometry. J Med Chem 49(1):205–214
Platania CBM, Salomone S, Leggio GM, Drago F, Bucolo C (2012) Homology modeling of dopamine D2 and D3 receptors: molecular dynamics refinement and docking evaluation. PLoS One 7(9):e44316
Platania CBM, Leggio GM, Drago F, Salomone S, Bucolo C (2013) Regulation of intraocular pressure in mice: structural analysis of dopaminergic and serotonergic systems in response to cabergoline. Biochem Pharmacol 86(9):1347–1356
Costanzi S (2013) Modeling G protein-coupled receptors and their interactions with ligands. Curr Opin Struct Biol 23(2):185–190
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31(13):3784–3788
Katritch V, Cherezov V, Stevens RC (2012) Diversity and modularity of G protein-coupled receptor structures. Trends Pharmacol Sci 33(1):17–27
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948. doi:10.1093/bioinformatics/btm404
Tusnady GE, Simon I (1998) Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J Mol Biol 283(2):489–506
Sonnhammer EL, von Heijne G, Krogh A (1998) A hidden Markov model for predicting transmembrane helices in protein sequences. Proc Int Conf Intell Syst Mol Biol 6:175-182
Cserzö M, Wallin E, Simon I, von Heijne G, Elofsson A (1997) Prediction of transmembrane alpha-helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng 10(6):673–676
Hirokawa T, Boon-Chieng S, Mitaku S (1998) SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14(4):378–379
Hofman K (1993) TMbase-A database of membrane spanning protein segments. Biol Chem Hoppe-Seyler 374:166
Käll L, Krogh A, Sonnhammer EL (2005) An HMM posterior decoder for sequence feature prediction that includes homology information. Bioinformatics 21(suppl 1):i251–i257
Rost B, Yachdav G, Liu J (2004) The predictprotein server. Nucleic Acids Res 32(suppl 2):W321–W326
Shokri A, Abedin A, Fattahi A, Kass SR (2012) J Am Chem Soc 134:10646–10650
Bernsel A, Viklund H, Hennerdal A, Elofsson A (2009) TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res: gkp363
Šali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234(3):779–815
Eswar N, Eramian D, Webb B, Shen MY, Sali A (2008) Protein structure modeling with MODELLER. Methods Mol Biol 426:145–159. doi:10.1007/978-1-60327-058-8_8
Zhang Y (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9(1):40
Ko J, Lee D, Park H, Coutsias EA, Lee J, Seok C (2011) The FALC-Loop web server for protein loop modeling. Nucleic Acids Res 39(suppl 2):W210–W214
Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT (2013) Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Res 41(W1):W349–W357
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26(2):283–291
Schmidt TH, Kandt C (2012) LAMBADA and InflateGRO2: efficient membrane alignment and insertion of membrane proteins for molecular dynamics simulations. J Chem Inf Model 52(10):2657–2669. doi:10.1021/ci3000453
Kandt C, Ash WL, Peter Tieleman D (2007) Setting up and running molecular dynamics simulations of membrane proteins. Methods 41(4):475–488
Allen WJ, Lemkul JA, Bevan DR (2009) GridMAT-MD: a grid-based membrane analysis tool for use with molecular dynamics. J Comput Chem 30(12):1952–1958. doi:10.1002/jcc.21172
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14(1):33–38
Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ (2005) GROMACS: fast, flexible, and free. J Comput Chem 26(16):1701–1718. doi:10.1002/jcc.20291
Laurie AT, Jackson RM (2005) Q-SiteFinder: an energy-based method for the prediction of protein–ligand binding sites. Bioinformatics 21(9):1908–1916
Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B (2012) ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res 40(D1):D1100–D1107
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) Open Babel: an open chemical toolbox. J Cheminformatics 3(1):1–14
Triballeau N, Acher F, Brabet I, Pin J-P, Bertrand H-O (2005) Virtual screening workflow development guided by the “receiver operating characteristic” curve approach. Application to high-throughput docking on metabotropic glutamate receptor subtype 4. J Med Chem 48(7):2534–2547
Sakhteman A, Lahtela-Kakkonen M, Poso A (2011) Studying the catechol binding cavity in comparative models of human dopamine D2 receptor. J Mol Graph Model 29(5):685–692. doi:10.1016/j.jmgm.2010.11.012
Vogel R, Mahalingam M, Lüdeke S, Huber T, Siebert F, Sakmar TP (2008) Functional role of the “ionic lock”—an interhelical hydrogen-bond network in family A heptahelical receptors. J Mol Biol 380(4):648–655
Kesten SR, Heffner TG, Johnson SJ, Pugsley TA, Wright JL, Wise LD (1999) Design, synthesis, and evaluation of chromen-2-ones as potent and selective human dopamine D4 antagonists. J Med Chem 42(18):3718–3725
Acknowledgments
The support from the Research Council at Shiraz University of Medical Sciences is acknowledged. The authors would like to thank Nazanin Bagherzadeh for her kind contribution in language editing of the manuscript. This work is a PharmD dissertation report, performed by Minasadat Khoddami, student of pharmacy at Shahid Sadoughi University of Medical Sciences.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Khoddami, M., Nadri, H., Moradi, A. et al. Homology modeling, molecular dynamic simulation, and docking based binding site analysis of human dopamine (D4) receptor. J Mol Model 21, 36 (2015). https://doi.org/10.1007/s00894-015-2579-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-015-2579-3