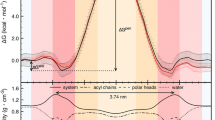
Abstract
Drug permeability determines the oral availability of drugs via cellular membranes. Poor permeability makes a drug unsuitable for further development. The permeability may be estimated as the free energy change that the drug should overcome through crossing membrane. In this paper the drug permeability was simulated using molecular dynamics method and the potential energy profile was calculated with potential of mean force (PMF) method. The membrane was simulated using DPPC bilayer and three drugs with different permeability were tested. PMF studies on these three drugs show that doxorubicin (low permeability) should pass higher free energy barrier from water to DPPC bilayer center while ibuprofen (high permeability) has a lower energy barrier. Our calculation indicates that the simulation model we built is suitable to predict drug permeability.





Similar content being viewed by others
References
Tian XJ, Yang XW, Yang X, Wang K (2009) Studies of intestinal permeability of 36 flavonoids using Caco-2 cell monolayer model. Int J Pharm 367:58–64
Hou TJ, Wang J, Zhang W, Wang W, Xu X (2006) Recent advances in computational prediction of drug absorption and permeability in drug discovery. Curr Med Chem 13:2653–2667
Kokate A, Li X, Williams PJ, Singh P, Jasti BR (2009) In silico prediction of drug permeability across buccal mucosa. Pharmaceut Res 26:1130–1139
Norinder U, Österberg T, Artursson P (1997) Theoretical calculation and prediction of Caco-2 cell permeability using Molsurf parametrization and PLS statistics. Pharmaceut Res 14:1786–1791
Acharya C, Seo PR, Polli JE, MacKerell AD Jr (2008) Computational model for predicting chemical substituent effects on passive drug permeability across parallel artificial membranes. Mol Pharmaceutics 5:818–828
Paixão P, Gouveia LF, Morais JAG (2010) prediction of the in vitro permeability determined in Caco-2 cells by using artificial neural networks. Eur J Pharm Sci 41:107–117
Rezai T, Bolk JE, Zhou MV, Kalyanaraman C, Lokey RS, Jacobson MP (2006) Conformational flexibility, internal hydrogen bonding, and passive membrane permeability. J Am Chem Soc 128:14073–14080
Xiang T, Xu Y, Anderson BD (1998) The barrier domain for solute permeation varies with lipid bilayer phase structure. J Membr Biol 165(1):77–90
Swift RV, Amaro RE (2011) Modeling the pharmacodynamics of passive membrane permeability. J Comput Aided Mol Des 25:1007–1017
Orsi M, Essex JW (2010) Permeability of drugs and hormones through a lipid bilayer. Soft Matter 6:3797–3808
Orsi M, Sanderson WE, Essex JW (2009) Permeability of small molecules through a lipid bilayer. J Phys Chem B 113:12019–12029
MacCallum JL, Bennett WFD, Tieleman DP (2008) Distribution of amino acids in a lipid bilayer from computer simulations. Biophys J 94:3393–3404
Boggara MB, Krishnamoorti R (2010) Partitioning of nonsteroidal anti-inflammatory drugs in lipid membranes. Biophys J 98:586–595
Hess B, Kutzner C, van der Spoel D, Lindahl E (2008) GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4:435–447
Berger O, Edholm O, Jähnig F (1997) Molecular dynamics simulations of a fluid bilayer of dipalmitoylphosphatidylcholine at full hydration, constant pressure and constant temperature. Biophys J 72:2002–2013
Tieleman DP, Berendsen HJC (1996) Molecular dynamics simulations of a fully hydrated dipalmitoylphosphatidylcholine bilayer with different macroscopic boundary conditions and parameters. J Chem Phys 105:4871–4880
Schuettelkopf AW, Aalten DMF (2004) PRODRG-a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D60:1355–1363
Hub JS, de Groot BL, van der Spoel D (2010) g_wham-A free weighted histogram analysis implementation including robust error and autocorrelation estimates. J Chem Theory Comput 6:3713–3720
Kyrychenko A, Sevriukov IY, Syzova ZA, Ladokhin AS, Doroshenko AO (2011) Partitioning of 2,6-Bis(1 H-Benzimidazol-2-yl)pyridine fluorophore into a phospholipid bilayer. Biophys Chem 154:8–17
Usansky HH, Sinko PJ (2005) Estimating human drug oral absorption kinetics from Caco-2 permeability using an absorption-disposition model. J Pharmacol Exp Ther 314:391–399
Paloncýová M, Berka K, Otyepka M (2012) Convergence of free energy profile of coumarin in lipid bilayer. J Chem Theory Comput 8:1200–1211
Kandasamy SK, Larson RG (2006) Molecular dynamics simulations of model trans-mambrane peptides in lipid bilayers. Biophys J 90:2326–2343
Acknowledgments
This work was supported by National Natural Science Foundation of China (No. 21103125) and China National Basic Research Program (No. 2010CB735602). MD simulations were performed on TianHe-1A supercomputer of National Supercomputing Center in Tianjin. The authors would like to thank the anonymous referees for valuable suggestions.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 340 kb)
Rights and permissions
About this article
Cite this article
Meng, F., Xu, W. Drug permeability prediction using PMF method. J Mol Model 19, 991–997 (2013). https://doi.org/10.1007/s00894-012-1655-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-012-1655-1