Abstract
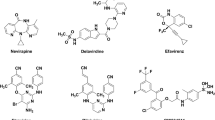
As a key component in combination therapy for acquired immunodeficiency syndrome (AIDS), non-nucleoside reverse transcriptase inhibitors (NNRTIs) have been proven to be an essential way in stopping HIV-1 replication. In the present work, in silico studies were conducted on a series of 119 NNRTIs, including 1-(2-hydroxyethoxymethyl)-6-(phenylthio)thymine (HEPT) and dihydroalkoxybenzyloxopyrimidine (DABO) derivatives by using the comparative molecular field analysis (CoMFA), comparative molecular similarity indices analysis (CoMSIA), docking simulations and molecular dynamics (MD). The statistical results of the optimal model, the ligand-based CoMSIA one (Q2 = 0.48, R 2ncv =0.847, R 2pre = 0.745) validates its satisfactory predictive capacity both internally and externally. The contour maps, docking and MD results correlate well with each other, drawing conclusions as follows: 1) Compounds with bulky substituents in position-6 of ring A, hydrophobic groups around position- 1, 2, 6 are preferable to the biological activities; 2) Two hydrogen bonds between RT inhibitor and the Tyr 318, Lys 101 residues, respectively, and a π-π bond between the inhibitor and Trp 188 are formed and crucial to the orientation of the active conformation of the molecules; 3) The binding pocket is essentially hydrophobic, which are determined by residues such as Trp 229, Tyr 318, Val 179, Tyr 188 and Val 108, and hydrophobic substituents may bring an improvement to the biological activity; 4) DABO and HEPT derivatives have different structures but take a similar mechanism to inhibit RT. The potency difference between two isomers in HEPTs can be explained by the distinct locations of the 6-naphthylmethyl substituent and the reasons are explained in details. All these results could be employed to alter the structural scaffold in order to develop new HIV-1 RT inhibitors that have an improved biological property. To the best of our knowledge, this is the first report on 3D-QSAR modeling of this series of HEPT and DABO NNRTs. The QSAR model and the information derived, we hope, will be of great help in presenting clear guidelines and accurate activity predictions for newly designed HIV-1 reverse transcriptase (RT) inhibitor.










Similar content being viewed by others
Reference
John PB, François P, Patrick G (2011) Global trends in AIDS mortality. In: Richard GR, Eileen MC (eds) The international handbook of adult mortality. Springer, New York, pp 171–183
Barre-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, Gruest J, Dauguet C, Axler-Blin C, Vezinet-Brun F, Rouzioux C, Rozenbaum W, Montagnier L (1983) Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220:868–871
Gallo RC, Sarin PS, Gelmann EP, Robert-Guroff M, Richardson E, Kalyanaraman VS, Mann D, Sidhu GD, Stahl RE, Zolla-Pazner S, Leibowitch J, Popovic M (1983) Isolation of human T-cell leukemia virus in acquired immune deficiency syndrome (AIDS). Science 220:865–867
Morris MC, Robert-Hebmann V, Chaloin L, Mery J, Heitz F, Devaux C, Goody RS, Divita G (1999) A new potent HIV-1 reverse transcriptase inhibitor. A synthetic peptide derived from the interface subunit domains. J Biol Chem 274:24941–24946
Gait MJ, Karn J (1995) Progress in anti-HIV structure-based drug design. Trends Biotechnol 13:430–438
Benbrik E, Chariot P, Bonavaud S, Ammi-SaId M, Frisdal E, Rey C, Gherardi R, Barlovatz-Meimon G (1997) Cellular and mitochondrial toxicity of zidovudine (AZT), didanosine (ddI) and zalcitabine (ddC) on cultured human muscle cells. J Neurol Sci 149:19–25
De Clercq E (1993) HIV-1-specific RT inhibitors: highly selective inhibitors of human immunodeficiency virus type 1 that are specifically targeted at the viral reverse transcriptase. Med Res Rev 13:229–258
Hopkins AL, Ren J, Esnouf RM, Willcox BE, Jones EY, Ross C, Miyasaka T, Walker RT, Tanaka H, Stammers DK, Stuart DI (1996) Complexes of HIV-1 reverse transcriptase with inhibitors of the HEPT series reveal conformational changes relevant to the design of potent non-nucleoside inhibitors. J Med Chem 39:1589–1600
Ren J, Esnouf R, Garman E, Somers D, Ross C, Kirby I, Keeling J, Darby G, Jones Y, Stuart D et al (1995) High resolution structures of HIV-1 RT from four RT-inhibitor complexes. Nat Struct Biol 2:293–302
Geretti AM (2006) Resistance to non-nucleoside reverse transcriptase inhibitors. Antiretroviral Resistance in Clinical Practice. Mediscript, London, Chapter 2
De Clercq E (2009) Anti-HIV drugs: 25 compounds approved within 25 years after the discovery of HIV. Int J Antimicrob Agents 33:307–320
De Clercq E (2009) The history of antiretrovirals: key discoveries over the past 25 years. Rev Med Virol 19:287
De Clercq E (2011) A 40-year journey in search of selective antiviral chemotherapy. Annu Rev Pharmacol Toxicol 51:1–24
Van de Waterbeemd H (1995) Introduction In Chemometric Methods in Drug Design. Chemometric Methods in Drug Design. VCH, Weinheim, chapter 1
Hao M, Li Y, Zhang S-W, Yang W (2011) Investigation on the binding mode of benzothiophene analogues as potent factor IXa (FIXa) inhibitors in thrombosis by CoMFA, docking and molecular dynamic studies. J Enzyme Inhib Med Chem. doi:10.3109/14756366.2011.554414
Liu J, Li Y, Zhang S, Xiao Z, Ai C (2011) Studies of new fused benzazepine as selective dopamine D3 receptor antagonists using 3D-QSAR, molecular docking and molecular dynamics. Int J Mol Sci 12:1196–1221
Zhang B, Li Y, Zhang H, Ai C (2010) 3D-QSAR and molecular docking studies on derivatives of MK-0457, GSK1070916 and SNS-314 as inhibitors against Aurora B kinase. Int J Mol Sci 11:4326–4347
Cramer RD, Patterson DE, Bunce JD (1988) Comparative molecular field analysis (CoMFA). 1. Effect of shape on binding of steroids to carrier proteins. J Am Chem Soc 110:5959–5967
Klebe G, Abraham U, Mietzner T (1994) Molecular similarity indices in a comparative analysis (CoMSIA) of drug molecules to correlate and predict their biological activity. J Med Chem 37:4130–4146
Baba M, Tanaka H, De Clercq E, Pauwels R, Balzarini J, Schols D, Nakashima H, Perno CF, Walker RT, Miyasaka T (1989) Highly specific inhibition of human immunodeficiency virus type 1 by a novel 6-substituted acyclouridine derivative. Biochem Biophys Res Commun 165:1375–1381
De Clercq E (1999) Perspectives of non-nucleoside reverse transcriptase inhibitors (NNRTIs) in the therapy of HIV-1 infection. II. Farmaco 54:26–45
Campiani G, Ramunno A, Maga G, Nacci V, Fattorusso C, Catalanotti B, Morelli E, Novellino E (2002) Non-nucleoside HIV-1 reverse transcriptase (RT) inhibitors: Past, present, and future perspectives (2002). Curr Pharm Design 8:615–657
Artico M, Massa S, Mai A, Marongiu EM, Piras G, Tramontano E, Colla LP (1993) 3,4-Dihydro-2-alkoxy-6-benzyl-4-oxopyrimidines (DABOs): a new class of specific inhibitors of human immunodeficiency virus Type 1. Antiviral chemistry & chemotherapy, vol 4. International Medical Press, London, pp 361–368
He Y, Chen F, Yu X, Wang Y, De Clercq E, Balzarini J, Pannecouque C (2004) Nonnucleoside HIV-1 reverse transcriptase inhibitors; part 3. Synthesis and antiviral activity of 5-alkyl-2-[(aryl and alkyloxyl-carbonylmethyl)thio]-6-(1-naphthylmethyl) pyrimidin-4(3H)-ones. Bioorg Chem 32:536–548
Ji L, Chen F-E, Feng X-Q, De Clercq E, Balzarini J, Pannecouque C (2006) Non-nucleoside HIV-1 reverse Transcriptase inhibitors, part 7. Synthesis, antiviral activity, and 3D-QSAR investigations of novel 6-(1-naphthoyl) HEPT analogues. Chem Pharm Bull 54:1248–1253
Meng G, Chen FE, De Clercq E, Balzarini J, Pannecouque C (2003) Nonnucleoside HIV-1 reverse transcriptase inhibitors: Part I. Synthesis and structure-activity relationship of 1-alkoxymethyl-5-alkyl-6-naphthylmethyl uracils as HEPT analogues. Chem Pharm Bull (Tokyo) 51:779–789
Sun GF, Chen XX, Chen FE, Wang YP, De Clercq E, Balzarini J, Pannecouque C (2005) Nonnucleoside HIV-1 reverse-transcriptase inhibitors, part 5. Synthesis and anti-HIV-1 activity of novel 6-naphthylthio HEPT analogues. Chem Pharm Bull (Tokyo) 53:886–892
Sun GF, Kuang YY, Chen FE, De Clercq E, Balzarini J, Pannecouque C (2005) Non-nucleoside HIV reverse transcriptase inhibitors, part 6[1]: synthesis and anti-HIV activity of novel 2-[(arylcarbonylmethyl)thio]-6-arylthio DABO analogues. Arch Pharm (Weinheim) 338:457–461
Wang Y, Chen FE, Balzarini J, De Clercq E, Pannecouque C (2008) Non-nucleoside HIV-1 reverse-transcriptase inhibitors. Part 10. Synthesis and anti-HIV activity of 5-alkyl-6-(1-naphthylmethyl)pyrimidin-4(3H)-ones with a mono- or disubstituted 2-amino function as novel 'dihydro-alkoxy-benzyl-oxopyrimidine' (DABO) analogues. Chem Biodivers 5:168–176
Gasteiger J, Marsili M (1980) Iterative partial equalization of orbital electronegativity–a rapid access to atomic charges. Tetrahedron 36:3219–3228
Zhu Y-Q, Lei M, Lu A-J, Zhao X, Yin X-J, Gao Q-Z (2009) 3D-QSAR studies of boron-containing dipeptides as proteasome inhibitors with CoMFA and CoMSIA methods. Eur J Med Chem 44:1486–1499
Deshpande S, Jaiswal S, Katti SB, Prabhakar YS (2011) CoMFA and CoMSIA analysis of tetrahydroquinolines as potential antimalarial agents. SAR QSAR Environ Res 1:1–16
Gerhard K (2002) Comparative molecular similarity indices analysis: CoMSIA. In: Hugo K, Gerd F, Yvonne CM (eds) 3D QSAR in Drug Design. Springer, New York, pp 87–104
Todeschini R, Consonni V, Mauri A, Pavan M (2004) DRAGON -Software for the calculation of molecular descriptors. Rel. 5.2 for Windows. Talete srl, Milano, Italy
Klebe G (1994) The use of composite crystal-field environments in molecular recognition and the de Novo design of protein ligands. J Mol Biol 237:212–235
Caballero J, Quiliano M, Alzate-Morales JH, Zimic M, Deharo E (2011) Docking and quantitative structure-activity relationship studies for 3-fluoro-4-(pyrrolo[2,1-f][1,2,4]triazin-4-yloxy)aniline, 3-fluoro-4-(1H-pyrrolo[2,3-b]pyridin-4-yloxy)aniline, and 4-(4-amino-2-fluorophenoxy)-2-pyridinylamine derivatives as c-Met kinase inhibitors. J Comput Aided Mol Des 25:349–369
Berendsen HJC, van der Spoel D, van Drunen R (1995) GROMACS: A message-passing parallel molecular dynamics implementation. Comput Phys Commun 91:43–56
Lindahl E, Hess B, Van der Spoel D (2001) GROMACS 3.0: a package for molecular simulation and trajectory analysis. J Mol Model 7:306–317
Aalten DMF, Bywater R, Findlay JBC, Hendlich M, Hooft RWW, Vriend G (1996) PRODRG, a program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules. J Comput Aided Mol Des 10:255–262
Schuttelkopf AW, van Aalten DMF (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr Sect D Biol Crystallogr 60:1355–1363
Berendsen HFC, Postma JPM, Van Gunsteren WF, Hermans J (1981) Interaction models for water in relation to protein hydration. In: Pullman B (ed) Inermolecular forces. Reidel, Dordrecht, pp 331–342
Parrinello M, Rahman A (1981) Polymorphic transitions in single crystals: A new molecular dynamics method. J Appl Phys 52:7182–7190
Lin JH, Perryman AL, Schames JR, McCammon JA (2002) Computational drug design accommodating receptor flexibility: The relaxed complex scheme. J Am Ceram Soc 124:5632–5633
Estrada E (1995) Edge adjacency relationships and a novel topological index related to molecular volume. J Chem Inf Comput Sci 35:31–33
Estrada E (1995) Edge adjacency relationships in molecular graphs containing heteroatoms: a new topological index related to molar volume. J Chem Inf Comput Sci 35:701–707
Meng G, Chen FE, De Clercq E (2003) Interactive study between two types of 1-[2-(Hydroxyethoxy)methyl]-6-naphthylmethylthymines and HIV-1 reverse transcriptase. Chin J Process Eng 3:24–28
Acknowledgments
This work is financially supported by the National Natural Science Foundation of China (Grants No. 10801025). The authors are also very grateful to Prof. Ling Yang for access to SYBYL software.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1–S10
(DOC 564 kb)
Rights and permissions
About this article
Cite this article
Mao, Y., Li, Y., Hao, M. et al. Docking, molecular dynamics and quantitative structure-activity relationship studies for HEPTs and DABOs as HIV-1 reverse transcriptase inhibitors. J Mol Model 18, 2185–2198 (2012). https://doi.org/10.1007/s00894-011-1236-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-011-1236-8