Abstract
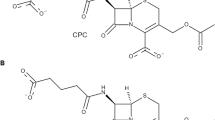
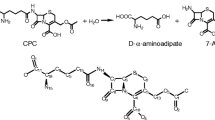
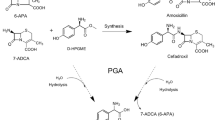
In the last step of penicillin biosynthesis, acyl-CoA:isopenicillin N acyltransferase (IAT) (E.C. 2.3.1.164) catalyzes the conversion of isopenicillin N (IPN) to penicillin G. IAT substitutes the α-aminoadipic acid side chain of IPN by a phenylacetic acid phenolate group (from phenylacetyl-CoA). Having a three-dimensional (3D) structure of IAT helps to determine the steps involved in side chain exchange by identifying the atomic details of substrate recognition. We predicted the IAT 3-D structure (α- and β-subunits), as well as the manner of IPN and phenylacetyl-CoA bind to the mature enzyme (β-subunit). The 3D IAT prediction was achieved by homology modeling and molecular docking in different snapshots, and refined by molecular dynamic simulations. Our model can reasonably interpret the results of a number of experiments, where key residues for IAT processing as well as strictly conserved residues most probably involved with enzymatic activity were mutated. Based on the results of docking studies, energies associated with the complexes, and binding constants calculated, we identified a site located in the region generated by β1, β2 and β5 strands, which forms part of the central structure of β-subunit, as the potential binding site of IPN. The site comprises the amino acid residues Cys103, Asp121, Phe122, Phe123, Ala168, Leu169, His170, Gln172, Phe212, Arg241, Leu262, Asp264, Arg302, Ser309, and Arg310. Through hydrogen bonds, the IPN binding site establishes interactions with Cys103, Leu169, Gln172, Asp264 and Arg310. Our model is also validated by a recently revealed crystal structure of the mature enzyme.














Similar content being viewed by others
References
Fawcett PA, Usher JJ, Abraham EP (1975) Behavior of tritium labeled isopenicillin N and 6-aminopenicillanic acid acyltransferase from Penicillium chrysogenum. Biochem J 151:741–746
Álvarez E, Cantoral JM, Barredo JL, Díez B, Martín JF (1987) Purification to homogeneity and characterization of acyl-coenzyme A:6-aminopenicillanic acid acyltransferase of Penicillium chrysogenum. Antimicrob Agents Chemother 31:1675–1682
Alonso MJ, Bermejo F, Reglero A, Fernández-Cañon JM, González de Buitrago G, Luengo JM (1988) Enzymatic síntesis of penicillins. J Antibiot 41:1074–1084
Whiteman PA, Abraham EP, Baldwin JE, Fleming MD, Schofield CJ, Sutherland JD, Willis AC (1990) Acyl-coenzyme A:6-aminopenicillanic acid acyltransferase from Penicillium chrysogenum and Aspergillus nidulans. FEBS Lett 262:342–344
Álvarez E, Meesschaert B, Montenegro E, Gutiérrez S, Díez B, Barredo JL, Martín JF (1993) The isopenicillin N acyltransferase of Penicillium chrysogenum has isopenicillin N amidohydrolase, 6-aminopenicillanic acid acyltransferase and penicillin amidase activities, all of wich are encoded by the single penDE gene. Eur J Biochem 215:323–332
Martin-Villacorta J, Reglero A, Luengo JM (1990) Acyl CoA: 6-APA acyltransferase from Penicillium chrysogenum. Studies on its hydrolitic activity. J Antibiot 44:108–110
Martin-Villacorta J, Reglero A, Luengo JM (1990) Biosynthesis of methoxy benzylpenicillins. Biotechnol Forum Europe 8:60–62
Ferrero MA, Reglero A, Martinez-Blanco H, Fernández-Valverde M, Luengo JM (1990) In vitro synthesis of new penicillins containing keto acids as side chains. Antimicrob Agents Chemother 35:1931–1932
Martínez-Blanco H, Reglero A, Luengo JM (1991) In vitro synthesis of different naturally-occurring, semisynthetic and synthetic penicillins using a new and effective enzymatic coupled system. J Antibiot 44:1252–1258
Barredo JL, Van Solingen P, Díez B, Álvarez E, Cantoral JM, Kattevilder A, Smaal EB, Groenen MAM, Veenstra AE, Martín JF (1989) Cloning and characterization of the acyl-coenzyme A:6-aminopenicillanic acid acyltransferase gene of Penicillium chrysogenum. Gene 83:291–300
Montenegro E, Barredo JL, Gutiérrez S, Díez B, Álvarez E, Martín JF (1990) Cloning, characterization of the acyl-CoA:6-aminopenicillanic acid acyltransferase gene of Aspergillus nidulans and linkage to the isopenicillin N synthase gene. Mol Gen Genet 221:322–330
Muller WH, Bovenberg RA, Groothuis MH, Kattevilder F, Smaal EB, Van der Voort LH, Verkleij AJ (1992) Involvement of microbodies in penicillin biosynthesis. Biochim Biophys Acta 1116:210–213
Muller WH, Essers J, Humbel BM, Verkleij AJ (1995) Enrichment of Penicillium chrysogenum microbodies by isopycnic centrifugation in nycodenz as visualized with immuno-electron microscopy. Biochim Biophys Acta 1245:215–220
Theilgaard HB, Kristiansen KN, Henriksen CM, Nielsen J (1997) Purification and characterization of delta-(l-alpha-aminoadipyl)-l-cysteinyl-d-valine synthetase from Penicillium chrysogenum. Biochem J 327:185–191
Aplin RT, Baldwin JE, Cole SCJ, Sutherland JD, Tobin MB (1993) On the production of α, β-heterodimeric acyl-coenzyme A:isopenicillin N acyltransferase of Penicillium chrysogenum: studies using a recombinant source. FEBS Lett 319:166–170
Aplin RT, Baldwin JE, Roach PL, Robinson CV, Schofiel CJ (1993) Investigations into the posttranslational modification and mechanism of isopenicillin N:acyl-CoA acyltransferase using electrospray mass spectrometry. Biochem J 294:357–363
Tobin MB, Baldwin JE, Cole SCJ, Miller JR, Skatrud PL, Sutherland JD (1993) The requeriment for subunit interation in the production of Penicillium crysogenum acyl-coenzyme A: isopenicillin N acyltransferase in Escherichia coli. Gene 132:199–206
Fernández FJ, Gutierrez S, Velasco J, Montenegro E, Marcos AT, Martín JF (1994) Molecular characterization of three loos-of-function mutations in the isopenicillin N-acyltransferase gene (pen DE) of Penicillium chrysogenum. J Bacteriol 176:4941–4948
Fernández FJ, Cardoza RE, Montenegro E, Velasco J, Gutiérrez S, Martín JF (2003) The isopenicillin N acyltransferases of Aspergillus nidulans and Penicillium chrysogenum differ in their ability to maintain the 40-kDa alpha beta heterodimer in an undissociated form. Eur J Biochem 270:1958–1968
Tobin MB, Cole SCJ, Kovacevic S, Miller JR, Baldwin JE, Sutherland JD (1994) Acyl-coenzyme A:isopenicillin N aciltranferase from Penicillium chrysogenum: effect of amino acid substitutions at Ser227, Ser230 and Ser309 on proenzyme cleavage and activity. FEMS Microbiol Lett 121:39–46
Tobin MB, Cole SCJ, Miller JR, Baldwin JE, Sutherland JD (1995) Amino-acid substitutions in the cleavage site of acyl-coenzyme A:isopenicillin N acyltransferase from Penicillium chrysogenum: effect on proenzyme cleavage and activity. Gene 162:29–35
Hensgens CM, Kroezinga EA, van Montfort BA, van der Laan JM, Sutherland JD, Dijkstra BW (2002) Purification, crystallization and preliminary X-ray diffraction of Cys103Ala acyl coenzyme A: isopenicillin N acyltransferase from Penicillium chrysogenum. Acta Crystallogr D58:716–718
Yoshida H, Hensgens CMH, van der Laan JM, Sutherland JD, Hart DJ, Dijkstra BW (2005) An approach to prevent aggregation during the purification and crystallization of wild type acyl coenzyme A:isopenicillin N acyltransferase from Penicillium chrysogenum. Protein Expr Purif 41:61–67
Bokhove M, Yoshida H, Hensgens CM, van der Laan JM, Sutherland JD, Dijkstra BW (2010) Structures of an isopenicillin N converting Ntn-hydrolase reveal different catalytic roles for the active site residues of precursor and mature enzyme. Structure 18:301–308
Mirky AE, Pauling L (1936) On the structure native, denatured and coagulated proteins. Proc Natl Acad Sci USA 22:439–447
Anfinsen CB, Haber E, Sela M, White FH (1961) The kinetics of formation of native ribonuclease during oxidation of the reduced polypeptide chain. Proc Natl Acad Sci USA 47:1309–1314
Havel TF, Snow ME (1991) A new method for building protein conformations from sequence alignments with homologues of known structure. J Mol Biol 217:1–7
Hilbert M, Bohm G, Jaenicke R (1993) Structural relationships of homologous proteins as a fundamental principle in homology modeling. Proteins 17:138–151
Sali A, Blundell TL (1993) Comparative protein modeling by satisfaction of spatial restraints. J Mol Biol 234:779–815
Srinivasan S, March CJ, Sudarsanam S (1993) An automated method for modeling proteins on known templates using distance geometry. Protein Sci 2:277–289
Bairoch A, Apweiler R (1997) The Swiss-Prot protein sequence database: its relevance to human molecular medical research. J Mol Med 75:312–316
Bennett-Lovsey RM, Herbert AD, Sternberg JE, Kelley LA (2008) Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins 70:611–625
Labarga A, Valentin F, Anderson M, Lopez R (2007) Web services at the European bioinformatics institute. Nucleic Acids Res 35:W6–W11
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Bowie JU, Lüthy R, Eisenberg D (1991) A method to identify protein sequences that fold into a known three-dimensional structure. Science 253:164–170
Flores TP, Orengo CA, Moss DS, Thornton JM (1993) Comparison of conformational characteristics in structurally similar protein pairs. Protein Sci 2:1811–1826
Raghava GPS (2002) APSSP2: A combination method for protein secondary structure prediction based on neural network and example based learning. CASP5 A–132
Rost B, Yachdav G, Liu J (2004) The PredictProtein Server. Nucleic Acids Res 32:W321–W326
Cole C, Barber JD, Barton GJ (2008) The Jpred 3 secondary structure prediction server. Nucleic Acids Res 36:W197–W201. doi:10.1093/nar/gkn238
Cuff JA, Clamp ME, Siddiqui AS, Finlay M, Barton GJ (1998) Jpred: a consensus secondary structure prediction server. Bioinformatics 14:892–893
Cuff JA, Barton GJ (1999) Evaluation and improvement of multiple sequence methods for protein secondary structure prediction. Protein Struct Funct Genet 34:508–519
Dong A, Xu X, Gu J, Edwards AM, Joachimiak A, Savchenko A (2007) Crystal structure of tetR-family transcriptional regulator. doi:10.2210/pdb2rek/pdb
Rossocha M, Schultz-Heienbrok R, von Moeller H, Coleman JP, Saenger W (2005) Conjugated bile acid hydrolase is a tetrameric N-terminal thiol hydrolase with specific recognition of its cholyl but not of its tauryl product. Biochemistry 44:5739–5748
Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31:3381–3385
Lambert C, Leonard N, De Bolle X, Depiereux E (2002) ESyPred3D: prediction of proteins 3D structures. Bioinformatics 18:1250–1256
Fiser A, Do RK, Sali A (2000) Modeling of loops in protein structures. Protein Sci 9:1753–1773
MacKerell AD, Bashford M, Bellot M, Dunbrack RL, Evanseck JD, Field MJ, Fisher S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus MJ (1998) All-atom empirical potencial for molecular modeling and dynamics studies of proteins. J Phys Chem B 102:3586–3616
Kale L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K, Schulten K (1999) NAMD2: greater scalability for parallel molecular dynamics. J Comput Phys 151:283–312
Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten KL (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26:1781–1802
Humphrey W, Dalke A, Schulten K (1996) VMD-Visual Molecular Dynamics. J Mol Graph 14:33–38
Diemand AV, Scheib H (2004) MolTalk, a programming library for protein structures and structure analysis. BMC Bioinformatics 5:1–7
Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, Snoeyink J, Richardson JS, Richardson DC (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35:W375–W383
Melo F, Feytmans E (1997) Novel knowledge-based mean force potential at atomic level. J Mol Biol 267:207–222
Melo F, Feytmans E (1998) Assessing protein structures with a non-local atomic interaction energy. J Mol Biol 277:1141–1152
Morris AL, MacArthur MW, Hutchinson EG, Thornton JM (1992) Stereochemical quality of protein structure coordinates. Proteins 12:345–364
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291
Leach AR (2001) Energy minimisation and related methods for exploring the energy surface. In Molecular modelling. Principles and applications. Prentice Hall, Englewood Cliffs, NJ
Tama F, Gadea FX, Marques O, Sanejouand YH (2000) Building-block approach for determining low-frequency normal modes of macromolecules. Proteins 41:1–7
Tama F, Sanejouand YH (2001) Conformational change of proteins arising from normal mode calculations. Protein Eng 14:1–6
Delarue M, Sanejouand YH (2002) Simplified normal mode analysis of conformational transitions in DNA-dependent polymerases: the elastic network model. J Mol Biol 320:1011–1024
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19:1639–1662
Hetényi C, Van der Spoel D (2002) Efficient docking of peptides to proteins without prior knowledge of the binding site. Protein Sci 11:1729–1737
Hetényi C, van der Spoel D (2006) Blind docking of drug-sized compounds to proteins with up to a thousand residues. FEBS Lett 580:1447–1450
Alfonso P, Pampín S, Estrada J, Rodríguez-Rey JC, Giraldo P, Sancho J, Pocoví M (2005) Miglustat (NB-DNJ) works as a chaperone for mutated acid beta-glucosidase in cells transfected with several Gaucher disease mutations. Blood Cells Mol Dis 35:268–276
Rosenfeld RJ, Goodsell DS, Musah RA, Morris GM, Goodin DB, Olson AJ (2003) Automated docking of ligands to an artificial active site: augmenting crystallographic analysis with computer modeling. J Comput Aided Mol Des 17:525–536
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Zakrzewski VG, Montgomery JA Jr, Stratmann RE, Burant JC, Dapprich S, Millam JM, Daniels AD, Kudin KN, Strain MC, Farkas O, Tomasi J, Barone V, Cossi M, Cammi R, Mennucci B, Pomelli C, Adamo C, Clifford S, Ochterski J, Peterson GA, Ayala P, Cui Q, Morokuma K, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Cioslowski J, Ortiz JV, Baboul AG, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Gomperts R, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Andres JL, Gonzalez C, Head-Gordon M, Replogle ES, Pople JA (1998) Gaussian 98, Revision A.9. Gaussian Inc, Pittsburgh, PA
Henikoff S, Henikoff JG (1992) Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci USA 89:10915–10919
Altschul SF (1991) Amino acid substitution matrices from an information theoretic perspective. J Mol Biol 219:555–565
Brannigan JA, Dodson G, Duggleby HJ, Moody PC, Smith JL, Tomchick DR, Murzin AG (1995) A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature 378:416–419
Pei J, Grishin NV (2003) Peptidase family U34 belongs to the superfamily of N-terminal nucleophile hydrolases. Protein Sci 12:1131–1135
Manavalan P, Curtis-Johnson W (1983) Sensitivity of circular dichroism to protein tertiary structure class. Nature 305:831–832
Liang J, Edelsbrunner H, Woodward C (1998) Anatomy of protein pockets and cavities: measurement of binding site geometry and implications for ligand design. Protein Sci 7:1884–1897
Liang J, Edelsbrunner H, Fu P, Sudhakar PV, Subramaniam S (1998) Analytical shape computation of macromolecules: II. Inaccessible cavities in proteins. Proteins 33:18–29
Kisselev OG, Kao J, Ponder JW, Fann YC, Gautam N, Marshall GR (1998) Light-activated rhodopsin induces structural binding motif in G protein alpha subunit. Proc Natl Acad Sci USA 95:4270–4275
Li HL, Galue A, Meadows L, Ragsdale DS (1999) A molecular basis for the different local anesthetic affinities of resting versus open and inactivated states of the sodium channel. Mol Pharmacol 55:134–141
Marshall GR, Ragno R, Makara GM, Arimoto R, Kisselev O (1999) Bound conformations for ligands for G-protein coupled receptors. Lett Pept Sci 6:283–288
Okada A, Miura T, Takeuchi H (2001) Protonation of histidine and histidine-tryptophan interaction in the activation of the M2 ion channel from influenza a virus. Biochemistry 40:6053–6060
Zacharias N, Dougherty DA (2002) Cation-pi interactions in ligand recognition and catalysis. Trends Pharmacol Sci 23:281–287
Quenner SW, Neuss N (1982) In: Morin EB, Morgan M (eds) The chemistry and biology of β-lactam antibiotics, vol 3. Academic, London, pp 1–81
Cremades N, Sancho J, Freire E (2006) The native-state ensemble of proteins provides clues for folding, misfolding and function. Trends Biochem Sci 31:494–496
Acknowledgments
The investigation was supported in part by grants from the Consejo Nacional de Ciencia y Tecnología (CONACyT), Mexico (Grant No. 132353), and Instituto Politécnico Nacional (Secretaría de Investigación y Posgrado and Comisión de Operación y Fomento de Actividades Académicas) and Instituto de Ciencia y Tecnología del Distrito Federal.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
More details on sequence alignment, homology modeling, molecular dynamics simulations and docking. This material and three-dimensional structures of IAT models described in this manuscript are available from the authors upon request.
Rights and permissions
About this article
Cite this article
Moreno-Vargas, L., Correa-Basurto, J., Maroun, R.C. et al. Homology modeling of the structure of acyl coA:isopenicillin N-acyltransferase (IAT) from Penicillium chrysogenum. IAT interaction studies with isopenicillin-N, combining molecular dynamics simulations and docking. J Mol Model 18, 1189–1205 (2012). https://doi.org/10.1007/s00894-011-1143-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-011-1143-z