Abstract
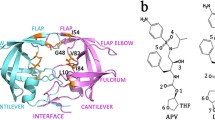
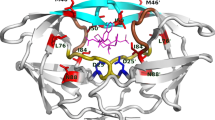
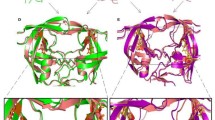
A major problem in the antiretroviral treatment of HIV-infections with protease-inhibitors is the emergence of resistance, resulting from the occurrence of distinct mutations within the protease molecule. In the present work we investigated the structural properties of a triple mutant (I54V-V82A-L90M) and a double mutant (V82A-L90M) that both confer strong resistance to ritonavir (RTV), but not to amprenavir (APV). For the unliganded double mutant protease molecular dynamics simulations revealed a contraction of the ligand binding pocket, which is enhanced by the I54V mutation. The observed displacement of backbone atoms of the 80s loops (residues 80–85 and 80’–85’ of the dimer) was found to primarily affect binding of the larger RTV molecule. The pocket contraction detected for the unbound protease upon mutation is also observed in the presence of APV, but not of RTV. As a consequence, the protein-ligand contacts lost upon the V82A mutation are restored by 80s loop motions for the APV-bound, but not for the RTV-bound form. RTV binding is therefore both hampered in the initial recognition step due to the poor fit of the bulky inhibitor into the small pocket of the mutant free protease and by the loss of protein-ligand interactions in the RTV-bound protease. The synergistic nature of both effects offers an explanation for the high level of resistance observed. These findings demonstrate that large inhibitors, which tightly bind to wild-type protease, may nevertheless be prone to the emergence of resistance in the presence of particular patterns of mutations. This information should be helpful for the design of novel and more effective drugs, e.g., by targeting different residues or by developing allosteric inhibitors that are capable of regulating protease dynamics.






Similar content being viewed by others
References
Mitsuya H, Yarchoan R, Broder S (1990) Molecular targets for AIDS therapy. Science 249:1533–1544
Kohl NE, Emmi EA, Schleif WA, Davies LJ, Heimbach JC, Dixon RAF (1988) Active human immunodeciency virus protease is required for viral infectivity. Proc Natl Acad Sci USA 85:686–690
Meek TD, Dayton BD, Metcalf BW, Dreyer GB, Strickler JE, Gorniak JG, Rosenberg M, Moore ML, Magaard VW, Debouck C (1989) Human immunodeficiency virus 1 protease expressed in Escherichia coli behaves as a dimeric aspartic protease. Proc Natl Acad Sci USA 86(6):1841–1845
Shafer RW (2006) Rationale and Uses of a Public HIV Drug-Resistance Database. J Infect Dis 194 Suppl 1:S51-8. Stanford HIV Database. http://hivdb.stanford.edu. Accessed 3 Dec 2009
Kanehisha Laboratories, Institute for Chemical Research, Kyoto University (2009) KEGG Release 52.1. http://www.genome.jp/kegg/drug. Accessed 3 Dec 2009
Kapoor A, Shapiro B, Shafer RW, Shulman N, Rhee SY, Delwart EL (2008) Multiple independent origins of a protease inhibitor resistance mutation in salvage therapy patients. Retrovirology 5:7
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank. Nucleic Acids Res 28(1):235–242
Kim EE, Baker CT, Dwyer MD, Murcko MA, Rao BG, Tung RD, Navia MA (1995) Crystal structure of HIV-1 protease in complex with VX-478, a potent and orally bioavailable Inhibitor of the Enzyme. J Am Chem Soc 117:1181–1182
Kempf DJ, Marsh KC, Denissen JF, McDonald E, Vasavanonda S, Flentge CA, Green BE, Fino L, Park CH, Kong XP et al. (1995) ABT-538 is a potent inhibitor of human immunodeficiency virus protease and has high oral bioavailability in humans. Proc Natl Acad Sci USA 92(7):2484–2488
Wartha F, Horn AHC, Meiselbach H, Sticht H (2005) Molecular dynamics simulations of HIV-1 protease suggest different mechanisms contributing to drug resistance. J Chem Theory Comput 1:315–324
Meiselbach H, Horn AH, Harrer T, Sticht H (2007) Insights into amprenavir resistance in E35D HIV-1 protease mutation from molecular dynamics and binding free-energy calculations. J Mol Model 13(2):297–304
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-Pdb Viewer: An environment for comparative protein modeling. Electrophoresis 18:2714–2723
Case DA, Pearlman DA, Caldwell JW III, TEC WJ, Ross WS, Simmerling CL, Darden TA, Merz KM, Stanton RV, Cheng AL, Vincent JJ, Crowley M, Tsui V, Gohlke H, Radmer RJ, Duan Y, Pitera J, Massova I, Seibel GL, Singh UC, Weiner PK, Kollman PA (2007) AMBER7. University of California, San Francisco
Pearlman DA, Case DA, Caldwell JW, Ross WS, Cheatham TE 3rd, DeBolt S, Ferguson D, Seibel G, Kollman P (1995) AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput Phys Commun 91:1–41
Case DA, Darden TA, Cheatham TE III, Simmerling CL, Wang J, Duke RE, Luo R, KMerz M, Pearlman DA, Crowley M, Walker RC, Zhang W, Wang B, Hayik S, Roitberg A, Seabra G, Wong KF, Paesani F, Wu X, Brozell S, Tsui V, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Beroza P, Mathews DH, Schafmeister C, Ross WS, Kollman PA (2006) AMBER 9. University of California, San Francisco
Cheatham TE, Cieplak P, Kollman PA (1999) A modified version of the Cornell et al. force field with improved sugar pucker phases and helical repeat. J Biomol Struct Dyn 16(4):845–862
Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KMJ, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA (1995) A second generation force field for the simulaton of proteins, nucleic acids and organic molecules. J Am Chem Soc 117:5179–5197
Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and testing of a general amber force field. J Comput Chem 25(9):1157–1174
Darden TA, York DM, Pedersen LG (1993) Particle mesh Ewald. An N.log(N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092
Ryckaert JP, Ciccotti G, Berendsen HJC (1977) Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Phys 23:327–341
SYBYL 7.3 (2008) Tripos International, 1699 South Hanley Rd., St. Louis, Missouri, 63144, USA
DS ViewerPro Suite. 6.0 (2005) Accelrys Software Inc., San Diego, CA
Hou T, Yu R (2007) Molecular dynamics and free energy studies on the wild-type and double mutant HIV-1 protease complexed with amprenavir and two amprenavir-related inhibitors: mechanism for binding and drug resistance. J Med Chem 50(6):1177–1188
Baldwin ET, Bhat TN, Liu B, Pattabiraman N, Erickson JW (1995) Structural basis of drug resistance for the V82A mutant of HIV-1 proteinase. Nat Struct Biol 2(3):244–249
Tie Y, Boross PI, Wang YF, Gaddis L, Liu F, Chen X, Tozser J, Harrison RW, Weber IT (2005) Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs. FEBS J 272(20):5265–5277
Liu F, Kovalevsky AY, Tie Y, Ghosh AK, Harrison RW, Weber IT (2008) Effect of flap mutations on structure of HIV-1 protease and inhibition by saquinavir and darunavir. J Mol Biol 381(1):102–115
Liu F, Kovalevsky AY, Louis JM, Boross PI, Wang YF, Harrison RW, Weber IT (2006) Mechanism of drug resistance revealed by the crystal structure of the unliganded HIV-1 protease with F53L mutation. J Mol Biol 358(5):1191–1199
Acknowledgments
This work was financially supported by the Johannes und Frieda Marohn-Stiftung. The authors also thank the Regionales Rechenzentrum Erlangen (RRZE) for technical support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Dirauf, P., Meiselbach, H. & Sticht, H. Effects of the V82A and I54V mutations on the dynamics and ligand binding properties of HIV-1 protease. J Mol Model 16, 1577–1583 (2010). https://doi.org/10.1007/s00894-010-0677-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-010-0677-9