Abstract
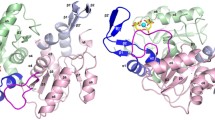
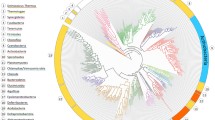
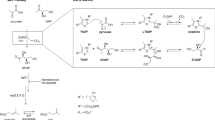
Tuberculosis is one of the leading infectious diseases in humans. Discovering new treatments for this disease is urgently required, especially in view of the emergence of multiple drug resistant organisms and to reduce the total duration of current treatments. The synthesis of isoprenoids in Mycobacterium tuberculosis has been reported as an interesting pathway to target, and particular attention has been focused on the methylerythritol phosphate (MEP) pathway comprising the early steps of isoprenoid biosynthesis. In this context we have studied the enzyme 2C-methyl-d-erythritol-4-phosphate cytidylyltransferase (CMS), the third enzyme in the MEP pathway, since the lack of a resolved structure of this protein in M. tuberculosis has seriously limited its use as a drug target. We performed homology modeling of M. tuberculosis CMS in order to provide a reliable model for use in structure-based drug design. After evaluating the quality of the model, we performed a thorough study of the catalytic site and the dimerization interface of the model, which suggested the most important sites (conserved and non-conserved) that could be useful for drug discovery and mutagenesis studies. We found that the metal coordination of CDP-methylerythritol in M. tuberculosis CMS differs substantially with respect to the Escherichia coli variant, consistent with the fact that the former is able to utilize several metal ions for catalysis. Moreover, we propose that electrostatic interactions could explain the higher affinity of the MEP substrate compared with the cytosine 5′-triphosphate substrate in the M. tuberculosis enzyme as reported previously.

Homology model of Mycobacterium tuberculosis CMS, for structure-based drug design of new antitubercular agents.











Similar content being viewed by others
References
Global tuberculosis control: surveillance, planning, financing (2007) (WHO/HTM/TB/2007376)
Vinícius M, de Souza N (2006) Promising drugs against tuberculosis. Recent Patents Anti-Infect Drug Disc 1:33–44
O’Brien RJ, Nunn PP (2001) The need for new drugs against tuberculosis. Obstacles, opportunities and next steps. Am J Respir Crit Care Med 162:1055–1058
Mukherjee JS, Rich ML, Socci AR, Joseph JK, Viru FA, Shin SS, Furin JJ, Becerra MC, Barry DJ, Kim JY, Bayona J, Farmer P, Fawzi MCS, Seung KJ (2004) Programs and principles in treatment of multidrug-resistant tuberculosis. Lancet 363:474–481
Wolucka BA, McNeil MR, Hoffmann E, Chojnacki T, Brennan PJ (1994) Recognition of the lipid intermediate for arabinogalactan/arabinomannan biosynthesis and its relation to the mode of action of ethambutol on mycobacteria. J Biol Chem 269:23328–23335
Mikusova K, Mikus M, Besra GS, Hancock I, Brennan PJ (1996) Biosynthesis of the linkage region of the mycobacteria cell wall. region. J Biol Chem 271:7820–7828
Mahapatra S, Yagi T, Belisle JT, Espinosa BJ, Hill PJ, McNeil MR, Brennan PJ, Crick DC (2005) Mycobacterial lipid II is composed of a complex mixture of modified muramyl and peptide moieties linked to decaprenyl phosphate. J Bacteriol 187:2747–2757
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol 100:221–230
Sacchettini JC, Poulter CD (1997) Creating isoprenoid diversity. Science 277:1788–1789
Bochar DA, Freisen JA, Stauffacher CV, Rodwell VW (1999) In: comprehensive natural products chemistry. Pergamon, Oxford
Testa CA, Brown MJ (2003) The methylerythritol phosphate pathway and its significance as a novel drug target. Curr Pharm Biotechnol 4:248–259
Rodríguez-Concepcion M (2004) The MEP pathway: a new target for the development of herbicides, antibiotics and antimalarial drugs. Curr Pharm Des 10:2391–2400
Eoh H, Brennan PJ, Crick DC (2008) The Mycobacterium tuberculosis MEP (2C-methyl-d-erythritol 4-phosphate) pathway as a new drug target. Tuberculosis 89:1–11
Sprenger GA, Schörken U, Wiegert T, Grolle S, De Graaf AA, Taylor SV, Begley TP, Bringer-Meyer S, Sahm H (1997) Identification of a thiamin-dependent synthase Escherichia coli required for the formation of 1-deoxy-d-xylulose 5-phosphate precursor to isoprenoids, thiamin and piridoxol. Proc Natl Acad Sci USA 94:12857–12862
Lois LM, Campos N, Putra SR, Danielsen K, Rohmer M, Boronat A (1998) Cloning and characterization of a gene from Escherichia coli encoding a transketolase-like enzyme that catalyzes the synthesis of d-1-deoxyxylulose 5-phosphate, a common precursor for isoprenoid, thiamin, and pyridoxol biosynthesis. Proc Natl Acad Sci USA 95:2105–2110
Lange BM, Wildung MR, McCaskill D, Croteau R (1998) A family of transketolases that directs isoprenoid biosynthesis via a mevalonate-independent pathway. Proc Natl Acad Sci USA 95:2100–2104
Kuzuyama T, Takahashi S, Watanabe H, Seto H (1998) Direct formation of 2-C-methyl-d-erythritol 4-phosphate from 1-deoxy-d-xylulose 5-phosphate by 1-deoxy-d-xylulose 5-phosphate reductoisomerase, a new enzyme in the Non-Mevalonate pathway to isopentenyl diphosphate. Tetrahedron Lett 39:4509–4512
Proteau PJ (2004) 1-Deoxy-d-xylulose 5-phosphate reductoisomerase: an overview. Bioorg Chem 32:483–493
Rohdich F, Wungsintaweekul J, Fellermeier M, Sagner S, Herz S, Kis K, Eisenreich W, Bacher A, Zenk MH (1996) Cytidine 5′-triphosphate-dependent biosynthesis of isoprenoids: YgbP protein of Escherichia coli catalyzes the formation of 4-diphosphocytidyl-2-C-methylerythritol. Proc Natl Acad Sci USA 96:11758–11763
Kuzuyama T, Takagi M, Kaneda K, Dairi T, Seto H (2000) Formation of 4-(cytidine 5′-diphospho)-2-C-methyl-d-erythritol from 2-C-methyl-d-erythritol 4-phosphate by 2-C-methyl-d-erythritol4-phosphate cytidylyltransferase, a new enzyme in the nonmevalonate pathway. Tetrahedron Lett 41:703–706
Richard SB, Bowman ME, Kwiatkowski W, Kang I, Chow C, Lillo AM, Cane DE, Noel JP (2001) Structure of 4-diphosphocytidyl-2-C-methylerythritol synthetase involved in mevalonate-independent isoprenoid biosynthesis. Nature Struct Biol 8:641–648
Richard S, Lillo A, Tetzlaff C, Bowman M, Noel J, Cane D (2004) Kinetic analysis of Escherichia coli 2-C-Methyl-d-erythritol-4-phosphate Cytidyltransferase, wild type and mutants, reveals roles of active site amino acids. Biochemistry 43:12189–12197
Cane DE, Chow C, Lillo A, Kang I (2001) Molecular cloning, expression and characterization of the first three genes in the mevalonate-independent isoprenoid pathway in Streptomyces coelicolor. Bioorg Med Chem 9:1467–1477
Shi W, Feng J, Zhang M, Lai X, Xu S, Zhang X, Wang H (2007) Biosynthesis of isoprenoids: characterization of a functionally active recombinant 2-C-methyl-d-erythritol 4-phosphate cytidyltransferase (IspD) from Mycobacterium tuberculosis H37Rv. Biochem Mol Biol 40:911–920
Eoh H, Brown AC, Buetow L, Hunter WH, Parish T, Kaur D, Brennan PJ, Crick DC (2007) Characterization of the Mycobacterium tuberculosis 4-diphosphocytidyl-2-C-methyl-d-erythritol synthase: potential for drug development. J Bacteriol 189:8922–8927
Chen L, Tsukuda M, Ebihara A, Shinkai A, Kuramitsu S, Yokoyama S, Chen LQ, Liu ZJ, Lee D, Chang SH, Nguyen D, Rose JP, Wang BC. Crystal structure of 2-C-methyl-d-erythritol 4-phosphate cytidylyltransferase from Thermus thermophilus HB8. Structure Summary MMDB, http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid=2PX7
Joint Center for Structural Genomics (JCSG). Joint Center for Structural Genomics (JCSG) Crystal structure of 2-C-methyl-d-erythritol 4-phosphate cytidylyltransferase (TM1393) from Thermotoga maritima at 2.67 A resolution. Structure Summary MMDB, http://www.ncbi.nlm.nih.gov/Structure/mmdb/mmdbsrv.cgi?uid=1VPA
Lillo AM, Tetzlaff CN, Sangari FJ, Cane DE (2003) Functional expression and characterization of EryA, the erythritol kinase of Brucella abortus, and enzymatic synthesis of l-erythritol-4-phosphate. Bioorg Med Chem Lett 13:737–739
Singh N, Avery MA, McCurdy CR (2007) Towards Mycobacterium tuberculosis DXR inhibitor design: homology modeling and molecular dynamics simulation. J Comput Aided Mol Des 21:511–522
Hillisch A, Pineda LF, Hilgenfeld R (2004) Utility of homology models in the drug discovery process. Drug Discov Today 9:659–669
Eswar N, Mari-Renom MA, Webb B, Madhusudhan MS, Eramian D, Shen M, Pieper U, Sali A (2006) Comparative protein structure modeling with MODELLER. Current protocols in Bioinformatics. Supplement 15, Wiley, New York, pp 5.6.1–5.6.30, 200
Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815
Fiser A, Do RK, Sali A (2000) Modeling of loops in protein structures. Protein Sci 9:1753–1773
Gabrielsen M, Bond CS, Hallyburton I, Hecht S, Bacher A, Einsenreich W, Rohdich F, Hunter WN (2004) Structure of the bifunctional ISPDF from Campylobacter jejuni. J Biol Chem 279:52753–52761
Patskovsky Y, Ho J, Toro R, Gilmore M, Miller S, Groshong C, Sauder MJ, Burley SK, Almo SC. Crystal structure of 2-C-methyl-d-erythritol-4-phosphate cytidyltransferase from Listeria monocytogenes.
Baur S, Marles-Wright J, Buckenmaier S, Lewis RJ, Vollmer W (2009) Synthesis of CDP-activated ribitol for teichoic acid precursors in Streptococcus pneumoniae. J Bacteriol 191:1200–1210
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612
Heinig M, Frishman D (2004) STRIDE: a web server for seconday structure assigment from known atomic coordinates of proteins. Nucleic Acids Res 32:W500–W502
Case DA, Pearlman DA, Caldwell JW, Cheathan TE III, Wang J, Ross WS, Simmerling CL, Darden TD, Merz KM, Stanton RV, Cheng AL, Vincent JJ, Crowley M, Tsui V, Gohlke H, Radmer RJ, Duan Y, Pitera J, Massova I, Seibel GL, Sligh UC, Weiner PK, Kollman PA (2002) AMBER 7. Univ California, San Francisco
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) Development and testing of a general amber force field. J Comput Chem 25:1157–1174
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Zakrzewski VG, Montgomery JA Jr, Stratmann RE, Burant JC, Dapprich S, Millam JM, Daniels AD, Kudin KN, Strain MC, Farkas O, Tomasi J, Barone V, Cossi M, Cammi R, Mennucci B, Pomelli C, Adamo C, Clifford S, Ochterski J, Petersson GA, Ayala PY, Cui Q, Morokuma K, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Cioslowski J, Ortiz JV, Baboul AG, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Gomperts R, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Gonzalez C, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Andres JL, Head-Gordon M, Replogle ES, Pople JA (1998) Gaussian 98. Revision A.6. Gaussian, Pittsburgh PA
Darden T, York D, Pedersen L (1993) Particle mesh Ewald: an W log(N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092
Cornell WD, Cieplak P, Bayly CI, Goud IR, Mertz KM Jr, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA (1995) A second generation of force fields for the simulation of proteins, nucleic acids and organic molecules. J Am Chem Soc 117:5179–5197
Berendsen HJC, Postman JPM, Van Gunsteren WF, DiNola A, Haak JA (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3690
Ryckaert JP, Ciccotti G, Berendsen HJ (1977) Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J Comput Chem 23:327–341
Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W, Donini O, Srivasan J, Case DA, Cheatam TE III (2000) Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res 33:889–897
Tsui V, Case DA (2001) Theory and applications of the generalized born solvation model in macromolecular simulations. Nucleic Acids Sci 56:275–291
Weiser J, Shemkin PS, Still WC (1999) Approximate atomic surfaces from linear combinations of pairwise overlaps. J Comput Chem 20:217–230
Pearlman DA (2005) Evaluating the molecular mechanics poisson-boltzmann surface area free energy method using a congeneric series of ligands to p38 MAP kinase. J Med Chem 48:7796–7807
Zang X, Li X, Wang R (2009) Interpretation of the binding affinities of PTP1B inhibitors with the MM-GB/SA method and the X-Score scoring function. J Chem Inf Model 49:1033–1048
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 26:283–291
Eisenberg D, Luthy R, Bowie JU (1997) VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol 277:396–404
Wiederstein M, Sippl MJ (2007) ProsaWeb: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35:407–410
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci 2:1511–1519
Hirsch AKH, Lauw S, Gersbach P, Schweizer WB, Rohdich F, Eisenreich W, Bacher A, Diederich F (2007) Nonphosphate inhibitors of IspE protein, a kinase in the non-mevalonate pathway for isoprenoid biosynthesis and a potential target for antimalarial therapy. Chemmedchem 2:806–810
Gohlke H, Kiel C, Case DA (2003) Insights into protein-protein binding by binding free energy calculations and free energy decomposition for Ras-Raf and Ras-RaIGDS complexes. Mol Biol 330:891–913
Obiol-Pardo C, Rubio-Martinez J (2008) Homology modeling of human transketolase: description of critical sites useful for drug design and study of the cofactor binding mode. J Mol Graph Model 27:723–734
Suhre K, Sanejouand YH (2004) ElNemo: a normal mode web-server for protein movement analysis and the generation of templates for molecular replacement. Nucleic Acids Res 32:W610–W614
Yaffe E, Fishelovitch D, Wolfson HJ, Halperin D, Nussinov (2008) MolAxis: efficient and accurate identification of channels in macromolecules. Prot Struct Funct Bioinform 73:72–86
Humprey W, Dalke A, Schulten K (1996) VMD visual molecular dynamics. J Mol Graph 14:33–38
Acknowledgments
The Spanish Ministry of Science and Technology supported this work through the projects CTQ2006-06588/BQU and BIO2002-04419-C02-02. The ‘Generalitat de Catalunya’ also supported this work through grants 2005SGR00914 and 2009SGR1308. We are also grateful to the ‘Departament d’Universitat, Recerca i Societat de la informació de la Generalitat de Catalunya i del Fons Social Europeu’.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Obiol-Pardo, C., Cordero, A., Rubio-Martinez, J. et al. Homology modeling of Mycobacterium tuberculosis 2C-methyl-d-erythritol-4-phosphate cytidylyltransferase, the third enzyme in the MEP pathway for isoprenoid biosynthesis. J Mol Model 16, 1061–1073 (2010). https://doi.org/10.1007/s00894-009-0615-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-009-0615-x