Abstract
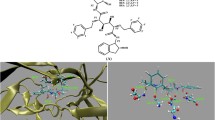
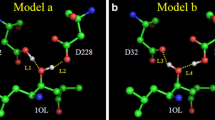
The molecular mechanics Poisson-Boltzmann surface area (MM-PBSA) method combined with molecular dynamics (MD) simulations were used to investigate the functional role of protonation in human immunodeficiency virus type 1 (HIV-1) protease complexed with the inhibitor BEA369. Our results demonstrate that protonation of two aspartic acids (Asp25/Asp25′) has a strong influence on the dynamics behavior of the complex, the binding free energy of BEA369, and inhibitor–residue interactions. Relative binding free energies calculated using the MM-PBSA method show that protonation of Asp25 results in the strongest binding of BEA369 to HIV-1 protease. Inhibitor–residue interactions computed by the theory of free energy decomposition also indicate that protonation of Asp25 has the most favorable effect on binding of BEA369. In addition, hydrogen-bond analysis based on the trajectories of the MD simulations shows that protonation of Asp25 strongly influences the water-mediated link of a conserved water molecule, Wat301. We expect that the results of this study will contribute significantly to binding calculations for BEA369, and to the design of high affinity inhibitors.







Similar content being viewed by others
References
Nam KY, Chang BH, Han CK, Ahn SK, No KT (2003) Bull Korean Chem Soc 6:817–823
Kohl NE, Emini EA, Schleif WA, Davis LI, Heimbach JC, Dixon RA, Scolnick EM, Sigal IS (1988) Proc Natl Acad Sci USA 85:4686–4690
Wlodawer A, Vondrasek J (1998) Annu Rev Biomol Struct 27:249–284
Smith R, Brereton IM, Chai RY, Kent SBH (1996) Nat Struct Biol 3:946–950
Chen X, Tropsha A (1995) J Med Chem 38:42–48
Wittayanarakul K, Aruksakunwong O, Saen-oon S, Chantratita W, Parasuk V, Sompornpisut P, Hannongbua S (2005) Biophys J 88:867–879
Yamazaki T, Nicholson LK, Torchia DA, Wingfield P, Stahl SJ, Kaufman JD, Eyermann CJ, Hodge CN, Lam PYS, Ru Y (1994) Am Chem Soc 116:10791–10792
Wang W, Kollman PA (2000) J Mol Biol 303:567–582
Andersson HO, Fridborg K, Lowgren S, Alterman M, Muhlman A, Bjorsne M, Garg N, Kvarnstrom I, Schaal W, Classon B, Karlen A, Danielsson UH, Ahlsen G, Nillroth U, Vrang L, Oberg B, Hallberg B, Samuelsson A, Unge T (2003) Eur J Biochem 270:1746–1758
Case DA, Darden TA, Cheatham III TE, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, Walker RC, Zhang W, Wang B, Hayik S, Roitberg A, Seabra G, Wong KF, Paesani F, Wu X, Brozell S, Tsui V, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Beroza P, Mathews DH, Schafmeister C, Ross WS, Kollman PA (2006) AMBER 9 University California, San Francisco
Cieplak P, Cornell WD, Bayly C, Kollman PA (1995) J Comput Chem 16:1357–1377
Ryckaert JP, Ciccotti G, Berendsen JC (1977) J Chem Phys 23:327–341
Darden T, York D, Pedersen L (1993) J Chem Phys 98:10089–10092
Li T, Matheus M, Herdewijn P (2008) J Mol Graphics Model 26:813–823
Hou T, Yu R (2007) J Med Chem 50:1177–1188
Xu Y, Wang R (2006) Proteins 64:1058–1068
Kuhn B, Gerber P, Schulz-Gasch T, Stahl M (2005) J Med Chem 48:4040–4048
Zhuang S, Zou J, Jiang Y, Mao X, Zhang B, Liu L, Yu Q (2005) J Med Chem 48:7208–7214
Wang J, Morin P, Wang W, Kollman PA (2001) J Am Chem Soc 123:5221–5230
Fogolari F, Brigo A, Molinari H (2003) Biophys J 85:159–166
Reyes CM, Kollman PA (2000) J Mol Biol 297:1145–1158
Sanner MF, Olson AJ, Spehner J (1996) Biopolymers 38:305–320
McQuarrie DA (1976) Statistical mechanics. Harper and Row, New York
Gohlke H, Kiel C, Case DA (2003) J Mol Biol 330:891–913
Yang R, Lee MC, Yan H, Duan Y (2005) Biophys J 89:95–106
Piana S, Carloni P, Rothlisberger U (2002) Protein Sci 11:2393–2402
Zoete V, Michielin O, Karplus M (2002) J Mol Biol 315:21–52
Lam PYS, Jahdav PK, Eyermann CJ, Hodge CN, Ru Y, Meek LT, Bacheler JL, Otto MJ, Rayner MM, Wong YN, Chang CH, Weber PC, Jackson DA, Sharpe TR, Erickson-Viitanen S (1994) Science 263:380–384
Lam PYS, Ru Y, Jahdav PK, Aldrich PE, DeLucca GV, Eyermann CJ, Chang CH, Emmett G, Holler ER, Daneker WF, Li L, Confalone PN, McHugh RJ, Han Q, Li R, Markwalder JA, Seitz SP, Sharpe TR, Bacheler LT, Rayner MM, Klabe RM, Shum L, Winslow DL, Korhauser DM, Jackson DA, Erickson-Viitanen S, Hodge CN (1996) J Med Chem 39:3514–3525
Hodge CN, Aldrich PE, Bacheler LT, Chang CH, Eyermann CJ, Garber S, Grubb M, Jackson DA, Jadhav PK, Korant B, Lam PY, Maurin MB, Meek JL, Otto MJ, Otto MM, Reid C, Sharpe TR, Shum L, Winslow DL, Erickson-Viitanen S (1996) Chem Biol 3:301–314
Lu Y, Yang CY, Wang S (2006) J Am Chem Soc 128:11830–11839
Davies MN, Toseland CP, Moss DM, Flower DR (2006) BMC Biochem 7:18 doi:10.1186/1471-2091-7-18
Acknowledgments
This work is supported by the National Nature Science Foundation of China (Grant Nos. 10874104, 10474060 and 10504017), the key Project of Chinese Ministry of Education (NO.206093) and the key Project of Nature Science Foundation of Shandong Province (Z2007A05).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, J., Yang, M., Hu, G. et al. Insights into the functional role of protonation states in the HIV-1 protease-BEA369 complex: molecular dynamics simulations and free energy calculations. J Mol Model 15, 1245–1252 (2009). https://doi.org/10.1007/s00894-009-0452-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-009-0452-y