Abstract
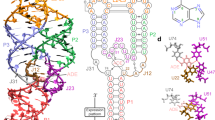
We present gas phase quantum chemical studies on the metabolite binding interactions in two important purine riboswitches, the adenine and guanine riboswitches, at the B3LYP/6-31G(d,p) level of theory. In order to gain insights into the strucutral basis of their discriminative abilities of regulating gene expression, the structural properties and binding energies for the gas phase optimized geometries of the metabolite bound binding pocket are analyzed and compared with their respective crystal geometries. Kitaura-Morokuma analysis has been carried out to calculate and decompose the interaction energy into various components. NBO and AIM analysis has been carried out to understand the strength and nature of binding of the individual aptamer bases with their respective purine metabolites. The Y74 base, U in case of adenine riboswitch and C in case of guanine riboswitch constitutes the only differentiating element between the two binding pockets. As expected, with W:W cis G:C74 interaction contributing more than 50% of the total binding energy, the interaction energy for metabolite binding as calculated for guanine (-46.43 Kcal/mol) is nearly double compared to the corresponding value for that of adenine (-24.73 Kcal/mol) in the crystal context. Variations in the optimized geometries for different models and comparison of relative contribution to metabolite binding involving four conserved bases reveal the possible role of U47:U51 W:H trans pair in the conformational transition of the riboswitch from the metabolite free to metabolite bound state. Our results are also indicative of significant contributions from stacking and magnesium ion interactions toward cooperativity effects in metabolite recognition.






Similar content being viewed by others
References
Gilbert SD, Stoddard CD, Wise SJ, Batey RT (2006) J Mol Biol 359:754–768
Nudler E, Mironov AS (2004) Trends Biochem Sci 29:11–17
Mandal M, Breaker RR (2004) Nat Rev Mol Cell Biol 5:451–463
Winkler W, Nahvi A, Breaker RR (2002) Nature 419:952–956
Mandal M, Boese B, Barrick JE, Winkler WC, Breaker RR (2003) Cell 113:577–586
Mandal M, Breaker RR (2004) Nat Struct Mol Biol 11:29–35
Grundy FJ, Lehman SC, Henkin TM (2003) Proc Natl Acad Sci USA 100:12057–12062
Sudarsan N, Wickiser JK, Nakamura S, Ebert MS, Breaker RR (2003) Genes Dev 17:2688–2697
Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL (1992) Ann Rev Phys Chem 43:257–282
Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR (2002) Chem Biol 9:1043–1049
Mironov A, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E (2002) Cell 111:747–756
Winkler W, Cohen-Chalamish S, Breaker RR (2002) Proc Natl Acad Sci USA 99:15908–15913
Winkler W, Nahvi A, Breaker RR (2002) Nature 419:952–956
McDaniel BAM, Grundy FJ, Artsimovitch I, Henkin TM (2003) Nat Struct Biol 10:701–707
Epshtein V, Mironov AS, Nudler E (2003) Proc Natl Acad Sci USA 100:3083–3088
Winkler W, Nahvi A, Sudarsan N, Barrick JE, Breaker RR (2003) Nat Struct Biol 10:701–707
Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I (2004) Proc Natl Acad Sci USA 101:6421–6426
Sudarsan N, Hammond MC, Block KL, Welz R, Barrick JE, Roth A, Breaker RR (2006) Science 314:300–304
Lea CR, Piccirilli JA (2007) Nat Chem Biol 3:16–17
Batey RT, Gilbert SD, Montage RK (2004) Nature 432:411–415
Ebbole DJ, Zalkin H (1987) J Biol Chem 262:8274–8287
Serganov A, Yuan Y, Pikovskaya O, Polonskaia A, Malinina I, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ (2004) Chem Biol 11:1729–1741
Mulhbacher J, Lafontaine DA (2007) Nucleic Acids Res 35:5568–5580
Lemay J-F, Lafontaine DA (2007) RNA 13:39–350
Ottink OM, Rampersad SM, Tssari M, Zaman GJR, Heus HA, Wijmenga SS (2007) RNA 13:2202–2212
manuscript under preparation
Gilbert SD, Mediatore SJ, Batey RT (2006) J Am Chem Soc 128:14214–14215
Gilbert SD, Love CE, Edwards AL, Batey RT (2007) Biochemistry 46:13297–13309
The Leontis and Westhof nomenclature for nucleic acids base pairing is employed using the following format: Base1:Base2 (A/U/G/C) Edge1:Edge2 (W/H/S) Glycosidic bond orienta- tion (Cis/Trans) where A, U, G, C, W, H and S stand for adenine, uracil, guanine, cytosine, Watson-Crick edge, Hoogsteen edge and sugar edge respectively. See Ref. [30] for details
Leontis NB, Westhof E (2001) RNA 7:499–512
Batey RT, Gilbert SD Chem Biol 13:805–807
Lemay J-F, Penedo JC, Tremblay R, Lilley DMJ, Lafontaine DA (2006) Chem Biol 13:857–868
Šponer J, Hobza P (2003) Collect Czech Chem Comm 68:2231–2282
Bhattacharyya D, Koripella SC, Mitra A, Rajendran VB, Sinha B (2007) J Biosci 32:809–825
Sharma P, Mitra A, Sharma S, Singh H (2007) J Chem Sci 119:525–531
Sharma P, Mitra A, Sharma S, Singh H, Bhattacharyya D (2008) J Biomol Struct Dyn 25:709–732
Sharma P, Singh H, Mitra A (2008) Lec Notes in Comp Sci 5102:379–386
Šponer JE, Spackova N, Leszczynski J, Šponer J (2005) J Phys Chem B 109:11399–11410
Šponer JE, Spackova N, Kulhanek P, Leszczynski J, Šponer J (2005) J Phys Chem A 109:2292–2301
Šponer JE, Spackova N, Sychrovsky’, Leszczynski J, Šponer J (2005) J Phys Chem B 109:18680–18689
Roy A, Panigrahi S, Bhattacharyya M, Bhattacharyya D (2008) J Phys Chem B 112:3786–3796
Šponer J, Jureˇcka, Hobza P (2004) J Am Chem Soc 126:10142–10151
Černý J, Hobza P (2007) Phys Chem Chem Phys 9:5291–5302
Kristyan S, Puly P (1994) Chem Phys Lett 229:175–180
Hobza P, Šponer J, Reschel T (1995) J Comput Chem 16:1315–1325
Hesselmann A, Jansen G, Schutz M (2005) J Chem Phys 122:014103
Misquitta AJ, Jeziorski B, Szalewicz K (2003) Phys Rev Lett 91:033201
Kohn W, Meir Y, Makarov DE (1998) Phys Rev Lett 80:4153–4156
Dion M, Rydberg H, Schroder E, Langreth DC, Lundqvist BI (2004) Phys Rev Lett 92:246401
Grimme S (2006) J Chem Phys 124:034108
Antony J, Grimme S (2006) Phys Chem Chem Phys 8:5287–5293
Grimme S (2006) J Comput Chem 27:1787–1799
Grimme S (2004) J Comput Chem 25:1463–1473
Morgado C, Vincent MA, Hillier IH, Shan X (2007) Phys Chem Chem Phys 9:448–451
Frisch MJ et al (2003) Gaussian03 revison B.05 Gaussian Inc Pittsburgh PA
Schmidt MW, Baldridge KK, Boatz JA, Elbert ST, Gordon MS, Jensen J, Koseky S, Matsunaga N, Nguyen KA, Su SJ, Windus TL, Dupius M, Montgomery JA (1993) J Comput Chem 14:1347–1363
Kitaura K, Morokuma K (1976) Int J Quantum Chem 10:325–331
Wang J, Gu J, Leszczynski (2005) J Phys Chem B 109:13761–13769
Wang J, Gu J, Leszczynski (2006) J Biomol Struct Dyn 24:139–148
Reed AE, Schleyer PvR (1987) J Am Chem Soc 109:7362–7373
Bader RFW (1990) Atoms in molecules. A quantum theory. The Clarendon Press, Oxford
Grimme S, Antony J, Schwabe T, Lichtenfeld CM (2007) 0rg Biomol Chem 5:741–758
Kolandaivel P, Nirmala V (2004) J Mol Struct 694:33–38
William S, Dudley H, Stephens E, O’Brien DP, Min Z (2004) Angew Chem 43:6596–6616
Acknowledgements
AM wishes to thank Department of Biotechnology (DBT), Government of India for research grant (NO. BT/PR5451/BID/07/111/2004. PS and SS thank the CSIR, New Delhi for research fellowships. Travel grants from DBT (for PS) and DST (for SS) for attending MDMM-2008, Piechwice, Poland are gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sharma, P., Sharma, S., Chawla, M. et al. Modeling the noncovalent interactions at the metabolite binding site in purine riboswitches. J Mol Model 15, 633–649 (2009). https://doi.org/10.1007/s00894-008-0384-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-008-0384-y