Abstract
High throughput docking (HTD) is routinely used for in silico screening of compound libraries with the aim to find novel leads in a drug discovery program. In the absence of an experimentally determined structure, a homology model can be used instead. Here we present an assessment of the utility of homology models in HTD by docking 300,000 anticipated inactive compounds along with 642 known actives into the binding site of the insulin-like growth factor 1 receptor (IGF-1R) kinase constructed by homology modeling. Twenty-one different templates were selected and the enrichment curves obtained by the homology models were compared to those obtained by three IGF-1R crystal structures. The results show a wide range of enrichments from random to as good as two of the three IGF-1R crystal structures. Nevertheless, if we consider the enrichment obtained at 2% of the database screened as a performance criterion, the best crystal structure outperforms the best homology model. Surprisingly, the sequence identity of the template to the target is not a good descriptor to predict the enrichment obtained by a homology model. The three homology models that yield the worst enrichment have the smallest binding-site volume. Based on our results, we propose ensemble docking to perform HTD with homology models.

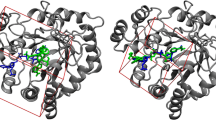
Top-scoring binding mode of NVP-AEW541 found by Glide with the aew receptor










Similar content being viewed by others
References
Jacobson MP, Sali A (2004) Ann Reports Med Chem 39:259–276
Hillisch A, Pineda LP, Hilgenfeld R (2004) Drug Discovery Today 9:659–669
Bissantz C, Bernard P, Hilbert M, Rognan D (2003) Proteins 50:5–25
Varady J, Wu X, Fang X, Min J, Hu Z, Levant B, Wang S (2003) J Med Chem 46:4377–4392
Evers A, Klabunde T (2005) J Med Chem 48:1088–1097
Salo OMH, Raitio KH, Savinainen JR, Nevalainen T, Lahtela-Kakkonen M, Laitinen JT, Järvinen T, Poso A (2005) J Med Chem 48:7166–7171
Vangrevelinghe E, Zimmermann K, Schoepfer J, Portmann R, Fabbro D, Furet P (2003) J Med Chem 46:2656–2662
McGovern SL, Shoichet BK (2003) J Med Chem 46:2895–2907
Diller DJ, Li R (2003) J Med Chem 46:4638–4647
Oshiro C, Bradley EK, Eksterowicz J, Evensen E, Lamb ML, Lanctot K, Putta S, Stanton R, Grootenhuis PDJ (2004) J Med Chem 47:764–767
Kairys V, Fernandes MX, Gilson MK (2006) J Chem Inf Model 46:365–379
Kenyon V, Chorny I, Carvajal WI, Holman TR, Jacobson MP (2006) J Med Chem 49:1356–1363
Blume-Jensen P, Hunter T (2001) Nature 411:355–365
Wagman AS, Nuss JM (2001) Curr Pharm Des 7:417–450
Adcock IM, Chung KF, Caramori G, Ito K (2006) Eur J Pharmacol 533:118–132
Verdonk ML, Berdini V, Hartshorn MJ, Mooij WTM, Murray CW, Taylor RD, Watson P (2004) J Chem Inf Comput Sci 44:793–806
Muegge I, Enyedy IJ (2004) Curr Med Chem 11:693–707
Chuaqui C, Deng Z, Singh J (2005) J Med Chem 48:121–133
Sims PA, Wong CF, McCammon JA (2003) J Med Chem:46:3314–3325
Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S (2002) Science 298:1912–1934
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) Nucleic Acids Research 28:235–242
Vieth M, Higgs RE, Robertson DH, Shapiro M, Gragg EA, Hemmerle H (2004) Biochim Biophys Acta 1697:243–257
Glide, version 4.0, Schrödinger, LLC, New York, NY, 2005
Garcia-Echeverria C, Pearson MA, Marti A, Meyer T, Mestan J, Zimmermann J, Gao JP, Brueggen J, Capraro HG, Cozens R, Evans DB, Fabbro D, Furet P, Porta DG, Liebetanz J, Martiny-Baron G, Ruetz S, Hofmann F (2004) Cancer Cell 5:231–239
Prime, version 1.5, Schrödinger, LLC, New York, NY, 2005
Maestro, version 7.5, Schrödinger, LLC, New York, NY, 2006
Sadowski J, Rudolph C, Gasteiger J (1990) Tetrahedron Comp Method 3:537–547
LigPrep, version 2.0, Schrödinger, LLC, New York, NY, 2005
Pipeline Pilot 5.0; Scitegic, Inc.
Witten IH, Frank E (1999) Data mining: practical machine learning tools and techniques with java implementations. Morgan Kaufmann Publishers, New York
Thomas MP, McInnes C, Fischer PM (2006) J Med Chem 49:92–104
Kleywegt GJ, Jones TA (1994) Acta Cryst D50:178–185
Ferrari AM, Wei BQ, Constantino L, Shoichet BK (2004) J Med Chem 47:5076–5084
Cavasotto CN, Abagyan RA (2004) J Mol Biol 337:209–225
Charifson PS, Corkery JJ, Murcko MA, Walters WP (1999) J Med Chem 42:5100–5109
Barril X, Morley SD (2005) J Med Chem 48:4432–4443
MacroModel XCluster, version 9.1, Schrödinger, LLC, New York, NY, 2005
Evers A, Gohlke H, Klebe G (2003) J Mol Biol 334:327–345
Acknowledgements
We thank Dr Nathan Brown for careful reading of the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ferrara, P., Jacoby, E. Evaluation of the utility of homology models in high throughput docking. J Mol Model 13, 897–905 (2007). https://doi.org/10.1007/s00894-007-0207-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-007-0207-6