Abstract
With the increasing amount of data provided by both high-throughput sequencing and structural genomics studies, there is a growing need for tools to augment functional predictions for protein sequences. Broad descriptions of function can be provided by establishing the presence of protein domains associated with a particular function. To extend the domain-based annotation, LigProf provides predictions of potential ligands that bind to a protein, as well as critical residues that stabilize ligands. A P-value statistic for estimating the significance of motif occurrence is provided for all sites. Although the usefulness of the method will rise with increasing numbers of crystallographically solved molecules deposited in the PDB database, we show that it can already be applied successfully to the highly represented ligand-bound protein kinase domains of viral and human origin. The LigProf webserver is freely available at: http://www.cropnet.pl/ligprof. At present, LigProf descriptors annotate and extend major protein families from the PfamA database.

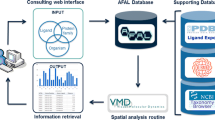
Information flow in LigProf database construction pipeline





Similar content being viewed by others
References
Hegyi H, Gerstein M (2001) Genome Res 11:1632–1640
Minshull J, Ness JE, Gustafsson C, Govindarajan S (2005) Curr Opin Chem Biol 9:202–209
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Nucl Acids Res 25:3389–3402
Devos D, Valencia A (2001) Trends Genet 17:429–431
Rost B, Liu J, Nair R, Wrzeszczynski KO, Ofran Y (2003) Cell Mol Life Sci 60:2637–2650
Green ML, Karp PD (2005) Nucl Acids Res 33:4035–4039
George RA, Spriggs RV, Thornton JM, Al-Lazikani B, Swindells MB (2004) Bioinformatics 20(Suppl 1):i130–i136
Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer EL, Bateman A (2006) Nucl Acids Res 34:D247–D251
Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, Ponting CP, Bork P (2006) Nucl Acids Res 32:D142–D144
Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWeese-Scott C, Geer LY, Gwadz M, He S, Hurwitz DI, Jackson JD, Ke Z, Lanczycki C, Liebert CA, Liu C, Lu F, Marchler GH, Mullokandov M, Shoemaker BA, Simonyan V, Song JS, Thiessen PA, Yamashita RA, Yin JJ, Zhang D, Bryant SH (2005) Nucl Acids Res 33:D192–196
George RA, Spriggs RV, Bartlett GJ, Gutteridge A, MacArthur MW, Porter CT, Al-Lazikani B, Thornton JM, Swindells MB (2005) Proc Natl Acad Sci USA 102:12299–12304
Gold ND, Jackson RM (2006) J Mol Biol 355:1112–1124
Shatsky M, Shulman-Peleg A, Nussinov R, Wolfson HJ (2006) J Comput Biol 13:407–428
Stoll V, Stewart KD, Maring CJ, Muchmore S, Giranda V, Gu YG, Wang G, Chen Y, Sun M, Zhao C, Kennedy AL, Madigan DL, Xu Y, Saldivar A, Kati W, Laver G, Sowin T, Sham HL, Greer J, Kempf D (2003) Biochemistry 42:718–727
Terasaka T, Kinoshita T, Kuno M, Seki N, Tanaka K, Nakanishi I (2004) J Med Chem 47:3730–3743
Schafferhans A, Klebe G (2001) J Mol Biol 307:407–427
Carpy AJ, Marchand-Geneste N (2006) SAR QSAR Environ Res 17:1–10
Snyder KA, Feldman HJ, Dumontier M, Salama JJ, Hogue CW (2006) BMC Bioinformatics 7:152
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) Nucl Acids Res 28:235–242
Sobolev V, Sorokine A, Prilusky J, Abola EE, Edelman M (1999) Bioinformatics 15:327–332
Eddy SR (1998) Bioinformatics 14:755–763
Pils B, Copley RR, Schultz J (2005) BMC Bioinformatics 6:210
Sneath PHA, Sokal RR (1973) WH Freeman, San Francisco
Gribskov M, McLachlan AD, Eisenberg D (1987) Proc Natl Acad Sci USA 84:4355–4358
Tudos E, Cserzo M, Simon I (1990) Int J Pept Protein Res 36:236–239
Magliery TJ, Regan L (2005) BMC Bioinformatics 30:240
Lichtarge O, Bourne HR, Cohen FE (1996) J Mol Biol 257:342–358
Mihalek I, Res I, Lichtarge O (2004) J Mol Biol 336:1265–1282
Johnson JM, Church GM (2000) Proc Natl Acad Sci USA 97:3965–3970
Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O’Donovan C, Phan I, Pilbout S, Schneider M (2003) Nucl Acids Res 31:365–370
Henikoff S, Henikoff JG (1992) Proc Natl Acad Sci USA 89:10915–10919
Wu TD, Nevill-Manning CG, Brutlag DL (2000) Bioinformatics 16:233–244
Sali A, Blundell TL (1993) J Mol Biol 234:779–815
Elofsson A (2002) Proteins 46:330–339
Kalinina OV, Mironov AA, Gelfand MS, Rakhmaninova AB (2004) Protein Sci 13:443–456
Hamelryck T, Manderick B (2003) Bioinformatics 19:2308–2310
Yang XL, Otero FJ, Skene RJ, McRee DE, Schimmel P, Pouplana L Ribas de (2003) Proc Natl Acad Sci USA 100:15376–15380
Li W, Jaroszewski L, Godzik A (2002) Bioinformatics 18:77–82
Qiu X, Janson CA, Smith WW, Green SM, McDevitt P, Johanson K, Carter P, Hibbs M, Lewis C, Chalker A, Fosberry A, Lalonde J, Berge J, Brown P, Houge-Frydrych CS, Jarvest RL (2001) Protein Sci 10:2008–2016
Hagglund R, Munger J, Poon AP, Roizman B (2002) J Virol 76:743–754
Kato A, Yamamoto M, Ohno T, Tanaka M, Sata T, Nishiyama Y, Kawaguchi Y (2006) J Virol 80:1476–1486
Prichard MN, Britt WJ, Daily SL, Hartline CB, Kern ER (2005) J Virol 79:15494–15502
Mestres J (2005) Drug Discov Today 10:1629–1637
Diwan P, Lacasse JJ, Schang LM (2004) J Virol 78:9352–9365
Schang LM, Coccaro E, Lacasse JJ (2005) Nucleosides Nucleotides Nucleic Acids 24:829–837
Guo T, Shi Y, Sun Z (2005) Prot Eng Des Sel 18:65–70
Shulman-Peleg A, Nussinov R, Wolfson HJ (2004) J Mol Biol 339:607–633
Schmitt S, Kuhn D, Klebe G (2002) J Mol Biol 323:387–406
Ondrechen MJ, Clifton JG, Ringe D (2001) Proc Natl Acad Sci USA 98:12473–12478
Kuhn D, Weskamp N, Schmitt S, Hullermeier E, Klebe G (2006) J Mol Biol 359:1023–1044
Ko J, Murga LF, Wei Y, Ondrechen MJ (2005) Bioinformatics 21:i258–i265
Kinoshita K, Nakamura H (2005) Protein Sci 14:711–718
Acknowledgements
This work has been supported by grants from the following EU projects: DATAGENOM (LSHB-CT-2003-503017) and GENEFUN (LSHG-CT-2004-503567). LSW was supported by a Program for Young Researchers from the Foundation of Polish Science and MNiSW research grants (2 P05A 001 30, PBZ-MNiI-2/1/2005).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Koczyk, G., Wyrwicz, L.S. & Rychlewski, L. LigProf: A simple tool for in silico prediction of ligand-binding sites. J Mol Model 13, 445–455 (2007). https://doi.org/10.1007/s00894-006-0165-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-006-0165-4