Abstract
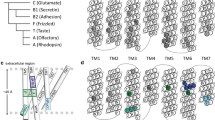
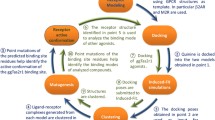
We employed the first principles computational method MembStruk and homology modeling techniques to predict the 3D structures of the human phenylthiocarbamide (PTC) taste receptor. This protein is a seven-transmembrane-domain G protein-coupled receptor that exists in two main forms worldwide, designated taster and nontaster, which differ from each other at three amino-acid positions. 3D models were generated with and without structural similarity comparison to bovine rhodopsin. We used computational tools (HierDock and ScanBindSite) to generate models of the receptor bound to PTC ligand to estimate binding sites and binding energies. In these models, PTC binds at a site distant from the variant amino acids, and PTC binding energy was equivalent for both the taster and nontaster forms of the protein. These models suggest that the inability of humans to taste PTC is due to a failure of G protein activation rather than decreased binding affinity of the receptor for PTC. Amino-acid substitutions in the sixth and seventh transmembrane domains of the nontaster form of the protein may produce increased steric hindrance between these two α-helices and reduce the motion of the sixth helix required for G protein activation.

3D-model of phenylthiocarbamide (PTC) bound to PTC taste reception







Similar content being viewed by others
References
Adler E, Hoon MA, Mueller KL, Chandrashekar J, Ryba NJ, Zuker CS (2000) Cell 100:93–702
Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, Guo W, Zuker CS, Ryba NJ (2000) Cell 100:703–711
Lindemann B (2001) Nature 413:219–225
Fox AL (1932) Proc Natl Acad Sci USA 18:115–120
Kim UK, Jorgenson E, Coon H, Leppert M, Risch N, Drayna D (2003) Science 299:1221–1225
Wooding S, Kim UK, Bamshad MJ, Larsen J, Jorde LB, Drayna D (2004) Am J Hum Genet 74:637–746
Fischer R, Griffin F, England S, Garn SM (1961) Nature 191:1328
Drewnowski A, Henderson SA, Barratt-Fornell A (2001) Drug Metab Dispos 29:535–538
Harris H, Kalmus H (1949) Ann Eugen 15:32–45
Barnicot N, Harris H, Kalmus H (1951) Ann Eugen 16:119–128
Delwiche JF, Buletic Z, Breslin PA (2001) Percept Psychophys 63:761–776
Kameswaran L, Gopalakrishnan S, Sukumar M (1974) Ind J Pharmac 6:134–140
Peterson DI, Lonergan LH, Hardinge MG (1968) Arch Environ Health 16:219–222
Cannon D, Baker T, Piper M, Scholand MB, Lawrence D, Drayna D, McMahon W, Villegas GM, Caton T, Coon H, Leppert M (2005) Nicotine Tob Res 7:853–858
Sultana T, Savage GP, Porter NG (2002) Proc Nutr Soc N Z 27:86–91
Floriano WB, Vaidehi N, Singer MS, Goddard WA III, Shepherd GM (2000) Proc Natl Acad Sci USA 97:10712–10716
Vaidehi N, Floriano WB, Trabanino R, Hall SE, Freddolino P, Choi EJ, Zamanakos G, Goddard WA III (2002) Proc Natl Acad Sci USA 99:12622–12627
Trabanino RJ, Hall S, Vaidehi N, Floriano WB, Goddard WA III (2004) Biophys J 84:1904–1921
Floriano WB, Nagarajan V, Zamanakos G, Goddard WA III (2004) J Med Chem 47:56–71
MDL Information Systems Inc, http://www.mdli.com
Tripos Inc, http://www.tripos.com
Gasteiger J, Marsili M (1980) Tetrahedron 36:3219–3228
Mayo SL, Olafson BD, Goddard WA III (1990) J Phys Chem 94:8897–8909
Accelrys Inc, http://www.accelrys.com
Vriend G (1990) J Mol Graph 8:52–56
MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiorkiewicz-Kuczera J, Yin D, Karplus M (1998) J Phys Chem B 102:3586–3616
Brady GP Jr, Stouten PFW (2000) J Comput Aided Mol Des 14:383–401
Freddolino PL, Yashar M, Kalani S, Vaidehi N, Floriano WB, Hall SE, Trabanino RJ, Kam VW, Goddard WA III (2004) Proc Natl Acad Sci USA 101:2736–2741
Floriano WB, Vaidehi N, Goddard WA III (2004) Chem Senses 29:269–290
Thompson JD, Higgins DG, Gibson TJ (1994) Nucleic Acids Res 22:4673–4680
Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RF, Yamamoto M, Miyano M (2000) Science 289:739–745
Hall MJ, Bartoshuk LM, Cain WS, Stevens JC (1975) Nature 253:442–443
Drewnowski A, Henderson SA, Shore AB (1997) Am J Clin Nutr 66:391–397
Bartoshuk LM (2000) Chem Senses 25:447–460
Farrens DL, Altenbach C, Yang K, Hubbell WL, Khorana HG (1996) Science 274:768–770
Sheikh SP, Zvyaga TA, Lichtarge O, Sakmar TP, Bourne HR (1996) Nature 383:347–350
Gether U, Lin S, Ghanouni P, Ballesteros JA, Weinstein H, Kobilka BK (1997) EMBO J 16:6737–6747
Dunham TD, Farrens DL (1999) J Biol Chem 274:1683–1690
Medkova M, Preininger AM, Yu NJ, Hubbell WL, Hamm HE (2002) Biochemistry 41:9962–9972
Schmidt C, Li B, Bloodworth L, Erlenbach I, Zeng FY, Wess J (2003) J Biol Chem 278:30248–30260
Pauwels PJ, Wurch T (1998) Mol Neurobiol 17:109–135
Gether U (2000) Endocr Rev 21:90–113
Okada T, Ernst OP, Palczewski K, Hofmann KP (2001) Trends Biochem Sci 26:318–324
Rao VR, Oprian DD (1996) Annu Rev Biophys Biomol Struct 25:287–314
Decaillot FM, Befort K, Filliol D, Yue S, Walker P, Kieffer BL (2003) Nat Struct Biol 10:629–636
Keast RS, Breslin PA (2002) Chem Senses 27:123–131
Schifferstein HNJ, Frijters JER (1991) Chem Senses 16:303–317
Acknowledgements
We thank Dr. Susan Sullivan and Dr. John Northup for helpful comments on the manuscript. This work was supported by NIDCD Z01-000046-04, and by NIH-BRGRO1-GM625523, NIH-R29AI40567, and NIH-HD36385. The computational facilities at the Materials and Process Simulation Center (MSC) were provided by a Shared University Research grant from International Business Machines and Defense University Research Instrumentation Program grants from the Army Research Office (ARO) and the Office of Naval Research (ONR). The facilities of the MSC are also supported by the Department of Energy-Advanced Simulation and Computing Program, National Science Foundation, Multidisciplinary Research Initiative - Army Research Office, Multidisciplinary Research Initiative - Office of Naval Research, General Motors, ChevronTexaco, Seiko-Epson, Beckman Institute, and Asahi Kasei.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Floriano, W.B., Hall, S., Vaidehi, N. et al. Modeling the human PTC bitter-taste receptor interactions with bitter tastants. J Mol Model 12, 931–941 (2006). https://doi.org/10.1007/s00894-006-0102-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-006-0102-6