Abstract
The molecular mechanisms of HIV drug resistance were studied using molecular dynamics simulations of HIV-1 protease complexes with the clinical inhibitor indinavir. One nanosecond molecular dynamics simulations were run for solvated complexes of indinavir with wild type protease, a control variant and 12 drug resistant mutants. The quality of the simulations was assessed by comparison with crystallographic and inhibition data. Molecular mechanisms that contribute to drug resistance include structural stability and affinity for inhibitor. The mutants showed a range of structural variation from 70 to 140% of the wild type protease. The protease affinity for indinavir was estimated by calculating the averaged molecular mechanics interaction energy. A correlation coefficient of 0.96 was obtained with observed inhibition constants for wild type and four mutants. Based on this good agreement, the trends in binding were predicted for the other mutants and discussed in relation to the clinical data for indinavir resistance.
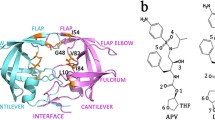
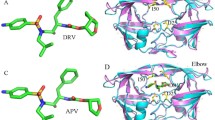
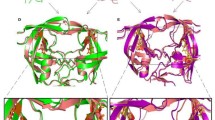
Figure Poincare map representation for WT protease-indinavir complex. The side chain of Tyr 59 showing the positions of hydrogen atoms.






Similar content being viewed by others
References
Tamalet C, Pasquier C, Yahi N, Colson P, Poizot-Martin I, Lepeu G, Gallais H, Massip P, Puel J, Izopet J (2000) J Med Virol 61:181–186
Shafer RW, Winters MA, Palmer S, Merigan TC (1998) Ann Intern Med 128:906–911
Hertogs K, Bloor S, Kemp SD, Van den Eynde C, Alcorn TM, Pauwels R, van Houtte M, Staszewski S, Miller V, Larder BA (2000) AIDS 14:1203–1210
Vergne L, Peeters M, Mpoudi-Ngole E, Bourgeois A, Liegeois F, Toure-Kane C, Mboup S, Mulanga-Kabeya C, Saman E, Jourdan J, Reynes J, Delaporte E (2000) J Clin Microbiol 38:3919–3925
Condra JH, Holder DJ, Schleif WA, Blahy OM, Danovich RM, Gabryelski LJ, Graham DJ, Laird D, Quintero JC, Rhodes A, Robbins HL, Roth E, Shivaprakash M, Yang T, Chodakewitz JA, Deutsch PJ, Leavitt RY, Massari FE, Mellors JW, Squires KE, Steigbigel RT, Teppler H, Emini EA (1996) J Virol 70:8270–8276
Brown AJ, Korber BT, Condra JH (1999) AIDS Res Hum Retroviruses 15:247–253
Rhee SY, Gonzales MJ, Kantor R, Betts BJ, Ravela J, Shafer RW (2003) Nucleic Acids Res 31:298–303
Shafer RW, Hsu P, Patick AK, Craig C, Brendel V (1999) J Virol 73:6197–6202
Shafer RW (2002) Clin Microbiol Rev 15:247–277
Wlodawer A, Vondrasek J (1998) Annu Rev Biophys Biomol Struct 27:249–284
Vacca JP, Dorsey BD, Schleif WA, Levin RB, McDaniel SL, Darke PL, Zugay J, Quintero JC, Blahy OM, Roth E, Sardana VV, Schlabach AJ, Graham PI, Condra JH, Gotlib L, Hollaway MK, Lin J, Chen I-W, Vastag K, Ostovic D, Anderson PS, Emini EA, Huff JR (1994) Proc Natl Acad Sci USA 91:4096–4100
Gulnik SV, Suvorov LI, Liu B, Yu B, Anderson B, Mitsuya H, Erickson JW (1995) Biochem 34:9282–9287
Chen Z, Li Y, Schock HB, Hall D, Chen E, Kuo LC (1995) J Biol Chem 270:433–436
King NM, Melnick L, Prabu-Jeyabalan M, Nalivaika EA, Yang SS, Gao Y, Nie X, Zepp C, Heefner DL, Schiffer CA (2002) Protein Sci 11:418–429
Muzammil S, Ross P, Freire E (2003) Biochemistry 42:631–638
Olsen DB, Stahlhut MW, Rutkowski CA, Schock HB, van Olden AL, Kuo LC (1999) J Biol Chem 274:23699–23701
Ohtaka H, Schon A, Freire E (2003) Biochemistry 42:13659–13666
Harrison RW, Weber IT (1994) Protein Eng 7:1353–1363
Liu H, Müller-Plathe F, van Gusteren WF (1996) J Mol Biol 261:454–469
Trylska J, Grochowski P, McCammon JA (2004) Protein Sci 13:513–528
Collins JR, Burt SK, Erickson JW (1995) Nat Struct Biol 2:334–338
Scott WR, Schiffer CA (2000) Structure Fold Des 8:1259–1265
Piana S, Carloni P, Rothlisberger U (2002) Protein Sci 11:2393–2402
Wang W, Kollman PA (2001) Proc Natl Acad Sci USA 98:14937–14942
Weber IT, Harrison RW (1999) Protein Eng 12:469–474
Mahalingam B, Louis JM, Reed CC, Adomat JM, Krouse J, Wang YF, Harrison RW, Weber IT (1999) Eur J Biochem 263:238–245
Harrison RW (1993) J Comp Chem 14:1112–1122
Weber IT, Harrison RW (1996) Protein Eng 9:679–690
Weber IT, Harrison RW (1997) Protein Sci 6:2365–2374
Bagossi P, Zahuczky G, Tozser J, Weber IT, Harrison RW (1999) J Mol Model 5:143–152
Harrison RW (1999) J Math Chem 26:125–137
Harrison RW (2003) Amortized fast multipole algorithm for molecular modeling. In: Dey PP, Amin MN, Gatton TM (eds) Proceedings of the International Conference on Computer Science and its Applications. pp 77–81
Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Acta Crystallogr A47:110–119
Sayle RA, Milner-White JE (1995) Trends Biochem Sci 20:374–376
Zoete V, Michielin O, Karplus M (2002) J Mol Biol 315:21–52
Chen Z, Li Y, Chen E, Hall DL, Darke PL, Culberson C, Shafer JA, Kuo LC (1994) J Biol Chem 269:26344–26348
Munshi S, Chen Z, Yan Y, Li Y, Olsen DB, Schock HB, Galvin BB, Dorsey B, Kuo LC (2000) Acta Crystallogr D56:381–388
Gustchina A, Weber IT (1990) FEBS Lett 269:269–272
Jenwitheesuk E, Samudrala R (2003) BMC Struct Biol 3:2
Acknowledgements
This work was supported in part by the Georgia Research Alliance and the United States Public Health Service Grants GM62920 and GM065762 (to I.T.W. and R.W.H.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, X., Weber, I.T. & Harrison, R.W. Molecular dynamics simulations of 14 HIV protease mutants in complexes with indinavir. J Mol Model 10, 373–381 (2004). https://doi.org/10.1007/s00894-004-0205-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00894-004-0205-x