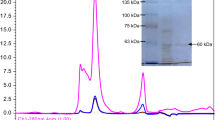
Abstract
The phrB gene encoding a putative cold-adapted DNA photolyase was cloned from the bacterial genomic DNA of Colwellia psychrerythraea 34H, a psychrophilic bacterium. Recombinant DNA photolyase, rCpPL, was overexpressed and purified from three different vectors. rCpPL binds its DNA substrate by flipping a cyclobutane pyrimidine dimer (CPD) into its active site and repairs CPD-containing DNA in vitro. rCpPL contains one catalytic flavin adenine dinucleotide (FAD) cofactor, but displays promiscuity in cofactor binding, in which either a flavin mononucleotide (FMN) or a methenyltetrahydrofolate (MTHF) molecule is bound as an antenna molecule and found in sub-stoichiometric amounts. The UV/Vis spectrum of oxidized rCpPL shows that the FADOX absorption maximum is the most red-shifted reported for a PL, suggesting a unique cavity electrostatic environment. Modest FAD vibronic structure suggests that the binding pocket is more flexible than warmer PLs, corroborating the hypothesis that psychrophilic proteins must be highly flexible to function at low temperatures. Fluorescence excitation data show that the freshly purified flavin cofactor is in its fully reduced state (FADH¯). A homology analysis of PL protein structures spanning 70 °C in growth temperature supports the data that the structure of CpPL is quite different from its warmer cousins.







Similar content being viewed by others
References
Albarracin VH et al (2014) First characterisation of a CPD-class I photolyase from a UV-resistant extremophile isolated from high-altitude Andean Lakes. Photochem Photobiol Sci 13:739–750. doi:10.1039/c3pp50399b
Altermark B, Niiranen L, Willassen NP, Smalas AO, Moe E (2007) Comparative studies of endonuclease I from cold-adapted Vibrio salmonicida and mesophilic Vibrio cholerae. FEBS J 274:252–263. doi:10.1111/j.1742-4658.2006.05580.x
Anderson S, Dragnea V, Masuda S, Ybe J, Moffat K, Bauer C (2005) Structure of a novel photoreceptor, the BLUF domain of AppA from Rhodobacter sphaeroides. Biochemistry 44:7998–8005
Artimo P et al (2012) ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res 40:W597–W603. doi:10.1093/nar/gks400
Aubert C, Mathis P, Eker AP, Brettel K (1999) Intraprotein electron transfer between tyrosine and tryptophan in DNA photolyase from Anacystis nidulans. Proc Natl Acad Sci USA 96:5423–5427
Bae E, Phillips GN (2004) Structures and analysis of highly homologous psychrophilic, mesophilic, and thermophilic adenylate kinases. J Biol Chem 279:28202–28208. doi:10.1074/jbc.M401865200
Benkert P, Kunzli M, Schwede T (2009) QMEAN server for protein model quality estimation. Nucleic Acids Res 37:W510–W514. doi:10.1093/nar/gkp322
Benkert P, Biasini M, Schwede T (2011) Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 27:343–350. doi:10.1093/bioinformatics/btq662
Berg BJV, Sancar GB (1998) Evidence for dinucleotide flipping by DNA photolyase. J Biol Chem 273:20276–20284
Biernat MA, Eker APM, van Oers MM, Vlak JM, van der Horst GTJ, Chaves I (2012) A baculovirus photolyase with DNA repair activity and circadian clock regulatory function. J Biol Rhythms 27:3–11. doi:10.1177/0748730411429665
Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31:3497–3500. doi:10.1093/nar/gkg500
Christine KS, MacFarlane AW IV, Yang K, Stanley RJ (2002) Cyclobutylpyrimidine dimer base flipping by DNA photolyase. J Biol Chem 277:38339–38344
Cipolla A, Delbrassine F, Da Lage JL, Feller G (2012) Temperature adaptations in psychrophilic, mesophilic and thermophilic chloride-dependent alpha-amylases. Biochimie 94:1943–1950. doi:10.1016/j.biochi.2012.05.013
Clivio P, Fourrey J-L (1996) (6-4) Photoproduct DNA photolyase mechanistic studies using s5-(6-4) photoproducts. Chem Commun 18:2203–2204
Costantini S, Colonna G, Facchiano AM (2008) ESBRI: a web server for evaluating salt bridges in proteins. Bioinformation 3:137–138
Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J (2006) CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res 34:W116–W118. doi:10.1093/nar/gkl282
Eker APM, Yajima H, Yasui A (1994) DNA photolyase from the fungus Neurospora crassa. Purification, characterization and comparison with other photolyases. Photochem Photobiol 60:125–133
Fujihashi M et al (2007) Crystal structure of archaeal photolyase from Sulfolobus tokodaii with two FAD molecules: implication of a novel light-harvesting cofactor. J Mol Biol 365:903–910
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31:3784–3788. doi:10.1093/nar/gkg563
Gauden M, van Stokkum IHM, Key JM, Luehrs DC, van Grondelle R, Hagemann P, Kennis TM (2006) Hydrogen-bond switching through a radical pair mechanism in a flavin-binding photoreceptor. Proc Natl Acad Sci USA 103:10895–10900
Gerday C et al (1997) Psychrophilic enzymes: a thermodynamic challenge. Biochim Biophys Acta (Protein Struct Mol Enzymol) 1342:119–131. doi:10.1016/s0167-4838(97)00093-9
Gindt YM, Edani BH, Olejnikova A, Roberts AN, Munshi S, Stanley RJ (2016) The missing electrostatic interactions between DNA substrate and Sulfolobus solfataricus DNA photolyase: what is the role of charged amino acids in thermophilic DNA binding proteins? J Phys Chem B 120:10234–10242. doi:10.1021/acs.jpcb.6b07201
Hamm-Alvarez S, Sancar A, Rajagopalan KV (1989) Role of enzyme-bound 5,10-methenyltetrahydropteroylpolyglutamate in catalysis by Escherichia coli DNA photolyase. J Biol Chem 264:9649–9656
Hopkins N, Stanley RJ (2003) Measurement of the electronic properties of the flavoprotein old yellow enzyme (OYE) and the OYE:p-Cl phenol charge-transfer complex using stark spectroscopy. Biochemistry 42:991–999
Hoyoux A et al (2004) Extreme catalysts from low-temperature environments. J Biosci Bioeng 98:317–330. doi:10.1263/jbb.98.317
Huang YH, Baxter R, Smith BS, Partch CL, Colbert CL, Deisenhofer J (2006) Crystal structure of cryptochrome 3 from Arabidopsis thaliana and its implications for photolyase activity. Proc Natl Acad Sci USA 103:17701–17706. doi:10.1073/pnas.0608554103
Islam SDM, Penzkofer A, Hegemann P (2003a) Quantum yield of triplet formation of riboflavin in aqueous solution and of flavin mononucleotide bound to the LOV1 domain of Photl from Chlamydomonas reinhardtii. Chem Phys 291:97–114. doi:10.1016/s0301-0104(03)00187-3
Islam SDM, Susdorf T, Penzkofer A, Hegemann P (2003b) Fluorescence quenching of flavin adenine dinucleotide in aqueous solution by pH dependent isomerisation and photo-induced electron transfer. Chem Phys 295:137–149. doi:10.1016/j.chemphys.2003.08.013
Johnson JL, Hamm-Alvarez S, Payne G, Sancar GB, Rajagopalan KV, Sancar A (1988) Identification of the second chromophore of Escherichia coli and yeast DNA photolyases as 5,10-methenyltetrahydrofolate. Proc Natl Acad Sci USA 85:2046–2050
Jorns MS, Wang B, Jordan SP, Chanderkar LP (1990) Chromophore function and interaction in Escherichia coli DNA photolyase: reconstitution of the apoenzyme with pterin and/or flavin derivatives. Biochemistry 29:552–561
Kao Y-T, Saxena C, Wang L, Sancar A, Zhong D (2005) Direct observation of thymine dimer repair in DNA by photolyase. Proc Natl Acad Sci USA 102:16128–16132
Kato R, Hasegawa K, Hidaka Y, Kuramitsu S, Hoshino T (1997) Characterization of a thermostable DNA photolyase from an extremely thermophilic bacterium, Thermus thermophilus HB27J. Bacteriology 179:6499–6503
Kay CWM, Bacher A, Fischer M, Richter G, Schleicher E, Weber S (2006) Blue light-initiated DNA repair by photolyase comprehensive series. Photochem Photobiol Sci 6:151–182
Kiener A, Husain I, Sancar A, Walsh C (1989) Purification and properties of Methanobacterium thermoautotrophicum DNA photolyase. J Biol Chem 264:13880–13887
Kim ST, Heelis PF, Okamura T, Hirata Y, Mataga N, Sancar A (1991) Determination of rates and yields of interchromophore (folate → flavin) energy transfer and intermolecular (flavin → DNA) electron transfer in Escherichia coli photolyase by time-resolved fluorescence and absorption spectroscopy. Biochemistry 30:11262–11270
Kim S-T, Malhotra K, Ryo H, Sancar A, Todo T (1996) Purification and characterization of Drosophila melanogaster photolyase. Mutat Res 363:97–104
Kim SY, Hwang KY, Kim SH, Sung HC, Han YS, Cho YJ (1999) Structural basis for cold adaptation—sequence, biochemical properties, and crystal structure of malate dehydrogenase from a psychrophile Aquaspirillium arcticum. J Biol Chem 274:11761–11767. doi:10.1074/jbc.274.17.11761
Kiontke S, Geisselbrecht Y, Pokorny R, Carell T, Batschauer A, Essen LO (2011) Crystal structures of an archaeal class II DNA photolyase and its complex with UV-damaged duplex DNA. EMBO J 30:4437–4449. doi:10.1038/emboj.2011.313
Klar T, Kaiser G, Hennecke U, Carell T, Batschauer A, Essen L-O (2006) Natural and non-natural antenna chromophores in the DNA photolyase from Thermus thermophilus. ChemBioChem 7:1798–1806
Kodali G, Siddiqui SU, Stanley RJ (2009) Charge redistribution in oxidized and semiquinone E. coli DNA photolyase upon photoexcitation: stark spectroscopy reveals a rationale for the position of Trp382. J Am Chem Soc 131:4795–4807
Komori H, Masui R, Kuramitsu S, Yokoyama S, Shibata T, Inoue Y, Miki K (2002) Crystal structure of thermostable DNA photolyase: pyrimidine-dimer recognition mechanism. Proc Natl Acad Sci USA 98:13560–13565
Kumar S, Nussinov R (2004) Different roles of electrostatics in heat and in cold: adaptation by citrate synthase. ChemBioChem 5:280–290. doi:10.1002/cbic.200300627
Langenbacher T, Zhao X, Bieser G, Heelis PF, Sancar A, Michel-Beyerle ME (1997) Substrate and temperature dependence of DNA photolyase repair activity examined with ultrafast spectroscopy. J Am Chem Soc 119:10532–10536
Leiros HKS, Pey AL, Innselset M, Moe E, Leiros I, Steen IH, Martinez A (2007) Structure of phenylalanine hydroxylase from Colwellia psychrerythraea 34H, a monomeric cold active enzyme with local flexibility around the active site and high overall stability. J Biol Chem 282:21973–21986. doi:10.1074/jbc.M610174200
Light DR, Walsh C, Marletta MA (1980) Analytical and preparative high-performance liquid chromatography separation of flavin and flavin analog coenzymes. Anal Biochem 109:87–93
Lipman RSA, Jorns MS (1992) Direct evidence for singlet-singlet energy transfer in Escherichia coli DNA photolyase. Biochemistry 31:786–791
Liptay W (1969) Electrochromism and Solvatochromism. Angew Chem Int Ed 8:177–188
Liu Z et al (2011) Dynamics and mechanism of cyclobutane pyrimidine dimer repair by DNA photolyase. Proc Natl Acad Sci. doi:10.1073/pnas.1110927108
Losi A, Gartner W (2012) The evolution of flavin-binding photoreceptors: an ancient chromophore serving trendy blue-light sensors. In: Merchant SS (ed) Annual review of plant biology, vol 63. Annual Reviews, Palo Alto, CA, USA, pp 49–72
MacFarlane AW IV, Stanley RJ (2001) Evidence of powerful substrate electric fields in DNA photolyase: implications for thymidine dimer repair. Biochemistry 40:15203–15214
MacFarlane AW IV, Stanley RJ (2003) Cis-syn thymidine dimer repair by DNA photolyase in real time. Biochemistry 42:8558–8568
Mees A, Klar T, Gnau P, Hennecke U, Eker APM, Carell T, Essen L-O (2004) Crystal structure of a photolyase bound to a CPD-like DNA lesion after in situ repair. Science 306:1789–1793
Methe BA et al (2005) The psychrophilic lifestyle as revealed by the genome sequence of Colwellia psychrerythraea 34H through genomic and proteomic analyses. Proc Natl Acad Sci USA 102:10913–10918
Miao LL et al (2016) Molecular structural basis for the cold adaptedness of the psychrophilic beta-glucosidase BglU in Micrococcus antarcticus. Appl Environ Microbiol 82:2021–2030. doi:10.1128/aem.03158-15
Nishimoto E, Aso Y, Koga T, Yamashita S (2006) Thermal unfolding process of dihydrolipoamide dehydrogenase studied by fluorescence spectroscopy. J Biochem 140:349–357. doi:10.1093/jb/mvj156
Ozturk N, Selby CP, Zhong D, Sancar A (2014) Mechanism of photosignaling by Drosophila cryptochrome role of the redox status of the flavin chromophore. J Biol Chem 289:4634–4642. doi:10.1074/jbc.M113.542498
Park H-W, Kim S-T, Sancar A, Deisenhofer J (1995) Crystal structure of DNA photolyase from Escherichia coli. Science 268:1866–1872
Parrilli E, Giuliani M, Marino G, Tutino ML (2010) Influence of production process design on inclusion bodies protein: the case of an Antarctic flavohemoglobin. Microb Cell Factories. doi:10.1186/1475-2859-9-19
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. doi:10.1002/jcc.20084
Sancar GB (1990) DNA photolyases: physical properties, action mechanism, and roles in dark repair. Mutat Res 236:147–160
Sancar A (2003) Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem Rev 103:2203–2237
Sancar A, Sancar GB (1984) Escherichia coli DNA photolyase is a flavoprotein. J Mol Biol 172:223–227
Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31:3381–3385. doi:10.1093/nar/gkg520
Selby CP, Sancar A (2006) A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc Natl Acad Sci USA 103:17696–17700
Selby CP, Sancar A (2012) The second chromophore in Drosophila photolyase/cryptochrome family photoreceptors. Biochemistry 51:167–171. doi:10.1021/bi201536w
Sheridan PP, Panasik N, Coombs JM, Brenchley JE (2000) Approaches for deciphering the structural basis of low temperature enzyme activity. Biochim Biophys Acta (Protein Struct Mol Enzymol) 1543:417–433. doi:10.1016/s0167-4838(00)00237-5
Siddiqui KS, Cavicchioli R (2006) Cold-adapted enzymes. Annu Rev Biochem 75:403–433. doi:10.1146/annurev.biochem.75.103004.142723
Soding J (2005) Protein homology detection by HMM–HMM comparison. Bioinformatics 21:951–960. doi:10.1093/bioinformatics/bti125
Stanley RJ (2001) Advances in flavin and flavoprotein optical spectroscopy. Antioxid Redox Signal 3:847–866
Stanley RJ, Siddiqui MS (2001) A Stark spectroscopic study of N(3)-methyl, N(10)-isobutyl-7,8-dimethylisoalloxazine in nonpolar low-temperature glasses: experiment and comparison with calculations. J Phys Chem A 105:11001–11008
Tamada T et al (1997) Crystal structure of DNA photolyase from Anacystis nidulans. Nat Struct Biol 4:887–891
Thiagarajan V, Byrdin M, Eker APM, Muller P, Brettel K (2011) Kinetics of cyclobutane thymine dimer splitting by DNA photolyase directly monitored in the UV. Proc Natl Acad Sci USA 108:9402–9407. doi:10.1073/pnas.1101026108
Thomas T, Cavicchioli R (1998) Archaeal cold-adapted proteins: structural and evolutionary analysis of the elongation factor 2 proteins from psychrophilic, mesophilic and thermophilic methanogens. FEBS Lett 439:281–286. doi:10.1016/s0014-5793(98)01375-1
Tina KG, Bhadra R, Srinivasan N (2007) PIC: protein interactions calculator. Nucleic Acids Res 35:W473–W476. doi:10.1093/nar/gkm423
Wang B, Jorns MS (1989) Reconstitution of Escherichia coli DNA photolyase with various folate derivatives. Biochemistry 28:1148–1152
Willey JM, Sherwood L, Woolverton C, Woolverton CJ (2008) Prescott, Harley, and Klein’s microbiology. McGraw-Hill, New York
Yang K, Stanley RJ (2006) Differential distortion of substrate occurs when it binds to DNA photolyase: a 2-aminopurine study. Biochemistry 45:11239–11245
Yang K, Matsika S, Stanley RJ (2007) 6MAP, a fluorescent adenine analogue, is a probe of base flipping by DNA photolyase. J Phys Chem B 111:10615–10625
Yasui A, Eker APM, Yasuhira S, Yajima H, Kobayashi T, Takao M, Oikawa A (1994) A new class of DNA photolyases present in various organisms including aplacental mammals. EMBO J 13:6143–6151
Acknowledgements
We wish to thank Mark Olsen and Cephalon for the donation of the Agilent HPLC. We also want to acknowledge Dr. Georges Feller, Dr. Aurora Martinez, Dr. Yvonne Gindt, and Dr. Peter S. Kessler for helpful discussions. The AnPL expression plasmid was a generous gift of Prof. A. Sancar, UNC Chapel Hill. S.M. and R.J.S. acknowledge support from NASA Exobiology Grant NNX13AH33G. This research was supported in part by the NSF Grant CHE-0847855. A.R. received support from NSF-REU supplement to CHE-0847855.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by L. Huang.
Electronic supplementary material
Below is the link to the electronic supplementary material.
792_2017_953_MOESM1_ESM.docx
Supplementary material 1 The SI includes buffer recipes, cloning, purification and harvesting protocols, photoreduction, and repair assay information. SI Figs. 1–11 include relative disorder probabilities and 3D homology structures for other PLs not discussed in the main text, HPLC analyses of cofactors and repair, absorption spectra, and SDS-PAGE analysis of the rCpPL-MBP fusion and rCpPL(S) system. Table SI-1 includes a calculation of FMN cofactor content (DOCX 11008 kb)
Rights and permissions
About this article
Cite this article
Munshi, S., Rajamoorthi, A. & Stanley, R.J. Characterization of a cold-adapted DNA photolyase from C. psychrerythraea 34H. Extremophiles 21, 919–932 (2017). https://doi.org/10.1007/s00792-017-0953-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-017-0953-z